vedo.pyplot
Advanced plotting functionalities.
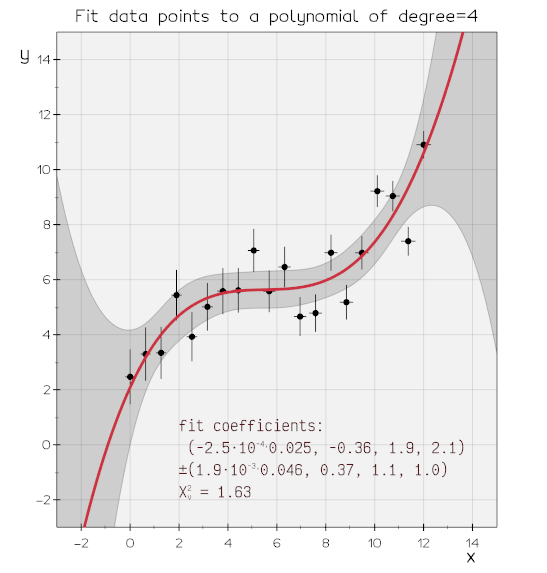
1#!/usr/bin/env python3 2# -*- coding: utf-8 -*- 3import warnings 4import numpy as np 5 6try: 7 import vedo.vtkclasses as vtk 8except ImportError: 9 import vtkmodules.all as vtk 10 11import vedo 12from vedo import settings 13from vedo import addons 14from vedo import colors 15from vedo import utils 16from vedo.pointcloud import merge 17from vedo.mesh import Mesh 18from vedo.assembly import Assembly, Group 19from vedo import shapes 20 21 22__docformat__ = "google" 23 24__doc__ = """ 25Advanced plotting functionalities. 26 27 28""" 29 30__all__ = [ 31 "Figure", 32 "Histogram1D", # uncomment to generate docs 33 "Histogram2D", 34 "PlotXY", 35 "PlotBars", 36 "plot", 37 "histogram", 38 "fit", 39 "donut", 40 "violin", 41 "whisker", 42 "streamplot", 43 "matrix", 44 "DirectedGraph", 45] 46 47########################################################################## 48def _to2d(actor, offset, scale): 49 50 poly = actor.polydata() 51 52 tp = vtk.vtkTransformPolyDataFilter() 53 transform = vtk.vtkTransform() 54 transform.Scale(scale, scale, scale) 55 transform.Translate(-offset[0], -offset[1], 0) 56 tp.SetTransform(transform) 57 tp.SetInputData(poly) 58 tp.Update() 59 60 poly = tp.GetOutput() 61 62 mapper2d = vtk.vtkPolyDataMapper2D() 63 mapper2d.SetInputData(poly) 64 65 act2d = vtk.vtkActor2D() 66 act2d.SetMapper(mapper2d) 67 68 act2d.GetProperty().SetColor(actor.color()) 69 act2d.GetProperty().SetOpacity(actor.alpha()) 70 act2d.GetProperty().SetLineWidth(actor.GetProperty().GetLineWidth()) 71 act2d.GetProperty().SetPointSize(actor.GetProperty().GetPointSize()) 72 73 act2d.PickableOff() 74 75 csys = act2d.GetPositionCoordinate() 76 csys.SetCoordinateSystem(4) 77 78 act2d.GetProperty().SetDisplayLocationToBackground() 79 return act2d 80 81 82########################################################################## 83class LabelData: 84 """Helper internal class to hold label information.""" 85 86 def __init__(self): 87 """Helper internal class to hold label information.""" 88 self.text = "dataset" 89 self.tcolor = "black" 90 self.marker = "s" 91 self.mcolor = "black" 92 93 94########################################################################## 95class Figure(Assembly): 96 """Format class for figures.""" 97 98 def __init__(self, xlim, ylim, aspect=4 / 3, padding=(0.05, 0.05, 0.05, 0.05), **kwargs): 99 """ 100 Create an empty formatted figure for plotting. 101 102 Arguments: 103 xlim : (list) 104 range of the x-axis as [x0, x1] 105 ylim : (list) 106 range of the y-axis as [y0, y1] 107 aspect : (float, str) 108 the desired aspect ratio of the histogram. Default is 4/3. 109 Use `aspect="equal"` to force the same units in x and y. 110 padding : (float, list) 111 keep a padding space from the axes (as a fraction of the axis size). 112 This can be a list of four numbers. 113 xtitle : (str) 114 title for the x-axis, can also be set using `axes=dict(xtitle="my x axis")` 115 ytitle : (str) 116 title for the y-axis, can also be set using `axes=dict(ytitle="my y axis")` 117 grid : (bool) 118 show the background grid for the axes, can also be set using `axes=dict(xygrid=True)` 119 axes : (dict) 120 an extra dictionary of options for the `vedo.addons.Axes` object 121 """ 122 123 self.verbose = True # printing to stdout on every mouse click 124 125 self.xlim = np.asarray(xlim) 126 self.ylim = np.asarray(ylim) 127 self.aspect = aspect 128 self.padding = padding 129 if not utils.is_sequence(self.padding): 130 self.padding = [self.padding, self.padding, self.padding, self.padding] 131 132 self.force_scaling_types = ( 133 shapes.Glyph, 134 shapes.Line, 135 shapes.Rectangle, 136 shapes.DashedLine, 137 shapes.Tube, 138 shapes.Ribbon, 139 shapes.GeoCircle, 140 shapes.Arc, 141 shapes.Grid, 142 # shapes.Arrows, # todo 143 # shapes.Arrows2D, # todo 144 shapes.Brace, # todo 145 ) 146 147 options = dict(kwargs) 148 149 self.title = options.pop("title", "") 150 self.xtitle = options.pop("xtitle", " ") 151 self.ytitle = options.pop("ytitle", " ") 152 number_of_divisions = 6 153 154 self.legend = None 155 self.labels = [] 156 self.label = options.pop("label", None) 157 if self.label: 158 self.labels = [self.label] 159 160 self.axopts = options.pop("axes", {}) 161 if isinstance(self.axopts, (bool, int, float)): 162 if self.axopts: 163 self.axopts = {} 164 if self.axopts or isinstance(self.axopts, dict): 165 number_of_divisions = self.axopts.pop("number_of_divisions", number_of_divisions) 166 167 self.axopts["xtitle"] = self.xtitle 168 self.axopts["ytitle"] = self.ytitle 169 170 if "xygrid" not in self.axopts: ## modify the default 171 self.axopts["xygrid"] = options.pop("grid", False) 172 173 if "xygrid_transparent" not in self.axopts: ## modify the default 174 self.axopts["xygrid_transparent"] = True 175 176 if "xtitle_position" not in self.axopts: ## modify the default 177 self.axopts["xtitle_position"] = 0.5 178 self.axopts["xtitle_justify"] = "top-center" 179 180 if "ytitle_position" not in self.axopts: ## modify the default 181 self.axopts["ytitle_position"] = 0.5 182 self.axopts["ytitle_justify"] = "bottom-center" 183 184 if self.label: 185 if "c" in self.axopts: 186 self.label.tcolor = self.axopts["c"] 187 188 x0, x1 = self.xlim 189 y0, y1 = self.ylim 190 dx = x1 - x0 191 dy = y1 - y0 192 x0lim, x1lim = (x0 - self.padding[0] * dx, x1 + self.padding[1] * dx) 193 y0lim, y1lim = (y0 - self.padding[2] * dy, y1 + self.padding[3] * dy) 194 dy = y1lim - y0lim 195 196 self.axes = None 197 if xlim[0] >= xlim[1] or ylim[0] >= ylim[1]: 198 vedo.logger.warning(f"Null range for Figure {self.title}... returning an empty Assembly.") 199 Assembly.__init__(self) 200 self.yscale = 0 201 return 202 203 if aspect == "equal": 204 self.aspect = dx / dy # so that yscale becomes 1 205 206 self.yscale = dx / dy / self.aspect 207 208 y0lim *= self.yscale 209 y1lim *= self.yscale 210 211 self.x0lim = x0lim 212 self.x1lim = x1lim 213 self.y0lim = y0lim 214 self.y1lim = y1lim 215 216 self.ztolerance = options.pop("ztolerance", None) 217 if self.ztolerance is None: 218 self.ztolerance = dx / 5000 219 220 ############## create axes 221 if self.axopts: 222 axes_opts = self.axopts 223 if self.axopts is True or self.axopts == 1: 224 axes_opts = {} 225 226 tp, ts = utils.make_ticks(y0lim / self.yscale, y1lim / self.yscale, number_of_divisions) 227 labs = [] 228 for i in range(1, len(tp) - 1): 229 ynew = utils.lin_interpolate(tp[i], [0, 1], [y0lim, y1lim]) 230 labs.append([ynew, ts[i]]) 231 232 if self.title: 233 axes_opts["htitle"] = self.title 234 axes_opts["y_values_and_labels"] = labs 235 axes_opts["xrange"] = (x0lim, x1lim) 236 axes_opts["yrange"] = (y0lim, y1lim) 237 axes_opts["zrange"] = (0, 0) 238 axes_opts["y_use_bounds"] = True 239 240 if "c" not in axes_opts and "ac" in options: 241 axes_opts["c"] = options["ac"] 242 243 self.axes = addons.Axes(**axes_opts) 244 245 Assembly.__init__(self, [self.axes]) 246 self.name = "Figure" 247 248 vedo.last_figure = self if settings.remember_last_figure_format else None 249 return 250 251 def _repr_html_(self): 252 """ 253 HTML representation of the Figure object for Jupyter Notebooks. 254 255 Returns: 256 HTML text with the image and some properties. 257 """ 258 import io 259 import base64 260 from PIL import Image 261 262 library_name = "vedo.pyplot.Figure" 263 help_url = "https://vedo.embl.es/docs/vedo/pyplot.html#Figure" 264 265 arr = self.thumbnail(zoom=1.1) 266 267 im = Image.fromarray(arr) 268 buffered = io.BytesIO() 269 im.save(buffered, format="PNG", quality=100) 270 encoded = base64.b64encode(buffered.getvalue()).decode("utf-8") 271 url = "data:image/png;base64," + encoded 272 image = f"<img src='{url}'></img>" 273 274 bounds = "<br/>".join( 275 [ 276 vedo.utils.precision(min_x, 4) + " ... " + vedo.utils.precision(max_x, 4) 277 for min_x, max_x in zip(self.bounds()[::2], self.bounds()[1::2]) 278 ] 279 ) 280 281 help_text = "" 282 if self.name: 283 help_text += f"<b> {self.name}:   </b>" 284 help_text += '<b><a href="' + help_url + '" target="_blank">' + library_name + "</a></b>" 285 if self.filename: 286 dots = "" 287 if len(self.filename) > 30: 288 dots = "..." 289 help_text += f"<br/><code><i>({dots}{self.filename[-30:]})</i></code>" 290 291 all = [ 292 "<table>", 293 "<tr>", 294 "<td>", 295 image, 296 "</td>", 297 "<td style='text-align: center; vertical-align: center;'><br/>", 298 help_text, 299 "<table>", 300 "<tr><td><b> nr. of parts </b></td><td>" + str(self.GetNumberOfPaths()) + "</td></tr>", 301 "<tr><td><b> position </b></td><td>" + str(self.GetPosition()) + "</td></tr>", 302 "<tr><td><b> x-limits </b></td><td>" + utils.precision(self.xlim, 4) + "</td></tr>", 303 "<tr><td><b> y-limits </b></td><td>" + utils.precision(self.ylim, 4) + "</td></tr>", 304 "<tr><td><b> world bounds </b> <br/> (x/y/z) </td><td>" + str(bounds) + "</td></tr>", 305 "</table>", 306 "</table>", 307 ] 308 return "\n".join(all) 309 310 def __add__(self, *obj): 311 # just to avoid confusion, supersede Assembly.__add__ 312 return self.__iadd__(*obj) 313 314 def __iadd__(self, *obj): 315 if len(obj) == 1 and isinstance(obj[0], Figure): 316 return self._check_unpack_and_insert(obj[0]) 317 318 obj = utils.flatten(obj) 319 return self.insert(*obj) 320 321 def _check_unpack_and_insert(self, fig): 322 323 if fig.label: 324 self.labels.append(fig.label) 325 326 if abs(self.yscale - fig.yscale) > 0.0001: 327 328 colors.printc(":bomb:ERROR: adding incompatible Figure. Y-scales are different:", 329 c='r', invert=True) 330 colors.printc(" first figure:", self.yscale, c='r') 331 colors.printc(" second figure:", fig.yscale, c='r') 332 333 colors.printc("One or more of these parameters can be the cause:", c="r") 334 if list(self.xlim) != list(fig.xlim): 335 colors.printc("xlim --------------------------------------------\n", 336 " first figure:", self.xlim, "\n", 337 " second figure:", fig.xlim, c='r') 338 if list(self.ylim) != list(fig.ylim): 339 colors.printc("ylim --------------------------------------------\n", 340 " first figure:", self.ylim, "\n", 341 " second figure:", fig.ylim, c='r') 342 if list(self.padding) != list(fig.padding): 343 colors.printc("padding -----------------------------------------\n", 344 " first figure:", self.padding, 345 " second figure:", fig.padding, c='r') 346 if self.aspect != fig.aspect: 347 colors.printc("aspect ------------------------------------------\n", 348 " first figure:", self.aspect, "\n", 349 " second figure:", fig.aspect, c='r') 350 351 colors.printc("\n:idea: Consider using fig2 = histogram(..., like=fig1)", c="r") 352 colors.printc(" Or fig += histogram(..., like=fig)\n", c="r") 353 return self 354 355 offset = self.zbounds()[1] + self.ztolerance 356 357 for ele in fig.unpack(): 358 if "Axes" in ele.name: 359 continue 360 ele.z(offset) 361 self.insert(ele, rescale=False) 362 363 return self 364 365 def insert(self, *objs, rescale=True, as3d=True, adjusted=False, cut=True): 366 """ 367 Insert objects into a Figure. 368 369 The recommended syntax is to use "+=", which calls `insert()` under the hood. 370 If a whole Figure is added with "+=", it is unpacked and its objects are added 371 one by one. 372 373 Arguments: 374 rescale : (bool) 375 rescale the y axis position while inserting the object. 376 as3d : (bool) 377 if True keep the aspect ratio of the 3d object, otherwise stretch it in y. 378 adjusted : (bool) 379 adjust the scaling according to the shortest axis 380 cut : (bool) 381 cut off the parts of the object which go beyond the axes frame. 382 """ 383 for a in objs: 384 385 if a in self.actors: 386 # should not add twice the same object in plot 387 continue 388 389 if isinstance(a, vedo.Points): # hacky way to identify Points 390 if a.ncells == a.npoints: 391 poly = a.polydata(False) 392 if poly.GetNumberOfPolys() == 0 and poly.GetNumberOfLines() == 0: 393 as3d = False 394 rescale = True 395 396 if isinstance(a, (shapes.Arrow, shapes.Arrow2D)): 397 # discard input Arrow and substitute it with a brand new one 398 # (because scaling would fatally distort the shape) 399 prop = a.GetProperty() 400 prop.LightingOff() 401 py = a.base[1] 402 a.top[1] = (a.top[1] - py) * self.yscale + py 403 b = shapes.Arrow2D(a.base, a.top, s=a.s, fill=a.fill).z(a.z()) 404 b.SetProperty(prop) 405 b.y(py * self.yscale) 406 a = b 407 408 # elif isinstance(a, shapes.Rectangle) and a.radius is not None: 409 # # discard input Rectangle and substitute it with a brand new one 410 # # (because scaling would fatally distort the shape of the corners) 411 # py = a.corner1[1] 412 # rx1,ry1,rz1 = a.corner1 413 # rx2,ry2,rz2 = a.corner2 414 # ry2 = (ry2-py) * self.yscale + py 415 # b = shapes.Rectangle([rx1,0,rz1], [rx2,ry2,rz2], radius=a.radius).z(a.z()) 416 # b.SetProperty(a.GetProperty()) 417 # b.y(py / self.yscale) 418 # a = b 419 420 else: 421 422 if rescale: 423 424 if not isinstance(a, Figure): 425 426 if as3d and not isinstance(a, self.force_scaling_types): 427 if adjusted: 428 scl = np.min([1, self.yscale]) 429 else: 430 scl = self.yscale 431 432 a.scale(scl) 433 434 else: 435 a.scale([1, self.yscale, 1]) 436 437 # shift it in y 438 a.y(a.y() * self.yscale) 439 440 if cut: 441 try: 442 bx0, bx1, by0, by1, _, _ = a.bounds() 443 if self.y0lim > by0: 444 a.cut_with_plane([0, self.y0lim, 0], [0, 1, 0]) 445 if self.y1lim < by1: 446 a.cut_with_plane([0, self.y1lim, 0], [0, -1, 0]) 447 if self.x0lim > bx0: 448 a.cut_with_plane([self.x0lim, 0, 0], [1, 0, 0]) 449 if self.x1lim < bx1: 450 a.cut_with_plane([self.x1lim, 0, 0], [-1, 0, 0]) 451 except: 452 # print("insert(): cannot cut", [a]) 453 pass 454 455 self.AddPart(a) 456 self.actors.append(a) 457 458 return self 459 460 def add_label(self, text, c=None, marker="", mc="black"): 461 """ 462 Manually add en entry label to the legend. 463 464 Arguments: 465 text : (str) 466 text string for the label. 467 c : (str) 468 color of the text 469 marker : (str), Mesh 470 a marker char or a Mesh object to be used as marker 471 mc : (str) 472 color for the marker 473 """ 474 newlabel = LabelData() 475 newlabel.text = text.replace("\n", " ") 476 newlabel.tcolor = c 477 newlabel.marker = marker 478 newlabel.mcolor = mc 479 self.labels.append(newlabel) 480 return self 481 482 def add_legend( 483 self, 484 pos="top-right", 485 relative=True, 486 font=None, 487 s=1, 488 c=None, 489 vspace=1.75, 490 padding=0.1, 491 radius=0, 492 alpha=1, 493 bc="k7", 494 lw=1, 495 lc="k4", 496 z=0, 497 ): 498 """ 499 Add existing labels to form a legend box. 500 Labels have been previously filled with eg: `plot(..., label="text")` 501 502 Arguments: 503 pos : (str, list) 504 A string or 2D coordinates. The default is "top-right". 505 relative : (bool) 506 control whether `pos` is absolute or relative, e.i. normalized 507 to the x and y ranges so that x and y in `pos=[x,y]` should be 508 both in the range [0,1]. 509 This flag is ignored if a string despcriptor is passed. 510 Default is True. 511 font : (str, int) 512 font name or number. 513 Check [available fonts here](https://vedo.embl.es/fonts). 514 s : (float) 515 global size of the legend 516 c : (str) 517 color of the text 518 vspace : (float) 519 vertical spacing of lines 520 padding : (float) 521 padding of the box as a fraction of the text size 522 radius : (float) 523 border radius of the box 524 alpha : (float) 525 opacity of the box. Values below 1 may cause poor rendering 526 because of antialiasing. 527 Use alpha = 0 to remove the box. 528 bc : (str) 529 box color 530 lw : (int) 531 border line width of the box in pixel units 532 lc : (int) 533 border line color of the box 534 z : (float) 535 set the zorder as z position (useful to avoid overlap) 536 """ 537 sx = self.x1lim - self.x0lim 538 s = s * sx / 55 # so that input can be about 1 539 540 ds = 0 541 texts = [] 542 mks = [] 543 for i, t in enumerate(self.labels): 544 label = self.labels[i] 545 t = label.text 546 547 if label.tcolor is not None: 548 c = label.tcolor 549 550 tx = vedo.shapes.Text3D(t, s=s, c=c, justify="center-left", font=font) 551 y0, y1 = tx.ybounds() 552 ds = max(y1 - y0, ds) 553 texts.append(tx) 554 555 mk = label.marker 556 if isinstance(mk, vedo.Points): 557 mk = mk.clone(deep=False).lighting("off") 558 cm = mk.center_of_mass() 559 ty0, ty1 = tx.ybounds() 560 oby0, oby1 = mk.ybounds() 561 mk.shift(-cm) 562 mk.SetOrigin(cm) 563 mk.scale((ty1 - ty0) / (oby1 - oby0)) 564 mk.scale([1.1, 1.1, 0.01]) 565 elif mk == "-": 566 mk = vedo.shapes.Marker(mk, s=s * 2) 567 mk.color(label.mcolor) 568 else: 569 mk = vedo.shapes.Marker(mk, s=s) 570 mk.color(label.mcolor) 571 mks.append(mk) 572 573 for i, tx in enumerate(texts): 574 tx.shift(0, -(i + 0) * ds * vspace) 575 576 for i, mk in enumerate(mks): 577 mk.shift(-ds * 1.75, -(i + 0) * ds * vspace, 0) 578 579 acts = texts + mks 580 581 aleg = Assembly(acts) # .show(axes=1).close() 582 x0, x1, y0, y1, _, _ = aleg.GetBounds() 583 584 if alpha: 585 dx = x1 - x0 586 dy = y1 - y0 587 588 if not utils.is_sequence(padding): 589 padding = [padding] * 4 590 padding = min(padding) 591 padding = min(padding * dx, padding * dy) 592 if len(self.labels) == 1: 593 padding *= 4 594 x0 -= padding 595 x1 += padding 596 y0 -= padding 597 y1 += padding 598 599 box = shapes.Rectangle([x0, y0], [x1, y1], radius=radius, c=bc, alpha=alpha) 600 box.shift(0, 0, -dy / 100).pickable(False) 601 if lc: 602 box.lc(lc).lw(lw) 603 aleg.AddPart(box) 604 605 xlim = self.xlim 606 ylim = self.ylim 607 if isinstance(pos, str): 608 px, py = 0, 0 609 rx, ry = (xlim[1] + xlim[0]) / 2, (ylim[1] + ylim[0]) / 2 610 shx, shy = 0, 0 611 if "top" in pos: 612 if "cent" in pos: 613 px, py = rx, ylim[1] 614 shx, shy = (x0 + x1) / 2, y1 615 elif "left" in pos: 616 px, py = xlim[0], ylim[1] 617 shx, shy = x0, y1 618 else: # "right" 619 px, py = xlim[1], ylim[1] 620 shx, shy = x1, y1 621 elif "bot" in pos: 622 if "left" in pos: 623 px, py = xlim[0], ylim[0] 624 shx, shy = x0, y0 625 elif "right" in pos: 626 px, py = xlim[1], ylim[0] 627 shx, shy = x1, y0 628 else: # "cent" 629 px, py = rx, ylim[0] 630 shx, shy = (x0 + x1) / 2, y0 631 elif "cent" in pos: 632 if "left" in pos: 633 px, py = xlim[0], ry 634 shx, shy = x0, (y0 + y1) / 2 635 elif "right" in pos: 636 px, py = xlim[1], ry 637 shx, shy = x1, (y0 + y1) / 2 638 else: 639 vedo.logger.error(f"in add_legend(), cannot understand {pos}") 640 raise RuntimeError 641 642 else: 643 644 if relative: 645 rx, ry = pos[0], pos[1] 646 px = (xlim[1] - xlim[0]) * rx + xlim[0] 647 py = (ylim[1] - ylim[0]) * ry + ylim[0] 648 z *= xlim[1] - xlim[0] 649 else: 650 px, py = pos[0], pos[1] 651 shx, shy = x0, y1 652 653 aleg.pos(px - shx, py * self.yscale - shy, self.z() + sx / 50 + z) 654 655 self.insert(aleg, rescale=False, cut=False) 656 self.legend = aleg 657 aleg.name = "Legend" 658 return self 659 660 def as2d(self, pos="bottom-left", scale=1, padding=0.05): 661 """ 662 Convert the Figure into a 2D static object (a 2D Assembly). 663 664 Arguments: 665 pos : (str, list) 666 position in 2D, as atring or list (x,y). 667 Any combination of "center", "top", "bottom", "left" and "right" will work. 668 The center of the renderer is [0,0] while top-right is [1,1]. 669 scale : (float) 670 scaling factor 671 padding : (float, list) 672 a single value or a list (xpad, ypad) 673 674 Returns: 675 `Group` object. 676 """ 677 x0, x1 = self.xbounds() 678 y0, y1 = self.ybounds() 679 pp = self.pos() 680 x0 -= pp[0] 681 x1 -= pp[0] 682 y0 -= pp[1] 683 y1 -= pp[1] 684 685 if not utils.is_sequence(padding): 686 padding = (padding, padding) 687 padding = np.array(padding) 688 689 if "cent" in pos: 690 offset = [(x0 + x1) / 2, (y0 + y1) / 2] 691 position = [0, 0] 692 if "right" in pos: 693 offset[0] = x1 694 position = [1 - padding[0], 0] 695 if "left" in pos: 696 offset[0] = x0 697 position = [-1 + padding[0], 0] 698 if "top" in pos: 699 offset[1] = y1 700 position = [0, 1 - padding[1]] 701 if "bottom" in pos: 702 offset[1] = y0 703 position = [0, -1 + padding[1]] 704 elif "top" in pos: 705 if "right" in pos: 706 offset = [x1, y1] 707 position = [1, 1] - padding 708 elif "left" in pos: 709 offset = [x0, y1] 710 position = [-1 + padding[0], 1 - padding[1]] 711 else: 712 raise ValueError(f"incomplete position pos='{pos}'") 713 elif "bottom" in pos: 714 if "right" in pos: 715 offset = [x1, y0] 716 position = [1 - padding[0], -1 + padding[1]] 717 elif "left" in pos: 718 offset = [x0, y0] 719 position = [-1, -1] + padding 720 else: 721 raise ValueError(f"incomplete position pos='{pos}'") 722 else: 723 offset = [0, 0] 724 position = pos 725 726 scanned = [] 727 group = Group() 728 for a in self.recursive_unpack(): 729 if a in scanned: 730 continue 731 if not isinstance(a, vedo.Points): 732 continue 733 if a.npoints == 0: 734 continue 735 if a.GetProperty().GetRepresentation() == 1: 736 # wireframe is not rendered correctly in 2d 737 continue 738 a2d = _to2d(a, offset, scale * 550 / (x1 - x0)) 739 a2d.SetPosition(position) 740 group += a2d 741 return group 742 743 744######################################################################################### 745class Histogram1D(Figure): 746 "1D histogramming." 747 748 def __init__( 749 self, 750 data, 751 weights=None, 752 bins=None, 753 errors=False, 754 density=False, 755 logscale=False, 756 fill=True, 757 radius=0.075, 758 c="olivedrab", 759 gap=0.02, 760 alpha=1, 761 outline=False, 762 lw=2, 763 lc="k", 764 texture="", 765 marker="", 766 ms=None, 767 mc=None, 768 ma=None, 769 # Figure and axes options: 770 like=None, 771 xlim=None, 772 ylim=(0, None), 773 aspect=4 / 3, 774 padding=(0.0, 0.0, 0.0, 0.05), 775 title="", 776 xtitle=" ", 777 ytitle=" ", 778 ac="k", 779 grid=False, 780 ztolerance=None, 781 label="", 782 **fig_kwargs, 783 ): 784 """ 785 Creates a `Histogram1D(Figure)` object. 786 787 Arguments: 788 weights : (list) 789 An array of weights, of the same shape as `data`. Each value in `data` 790 only contributes its associated weight towards the bin count (instead of 1). 791 bins : (int) 792 number of bins 793 density : (bool) 794 normalize the area to 1 by dividing by the nr of entries and bin size 795 logscale : (bool) 796 use logscale on y-axis 797 fill : (bool) 798 fill bars with solid color `c` 799 gap : (float) 800 leave a small space btw bars 801 radius : (float) 802 border radius of the top of the histogram bar. Default value is 0.1. 803 texture : (str) 804 url or path to an image to be used as texture for the bin 805 outline : (bool) 806 show outline of the bins 807 errors : (bool) 808 show error bars 809 xtitle : (str) 810 title for the x-axis, can also be set using `axes=dict(xtitle="my x axis")` 811 ytitle : (str) 812 title for the y-axis, can also be set using `axes=dict(ytitle="my y axis")` 813 padding : (float), list 814 keep a padding space from the axes (as a fraction of the axis size). 815 This can be a list of four numbers. 816 aspect : (float) 817 the desired aspect ratio of the histogram. Default is 4/3. 818 grid : (bool) 819 show the background grid for the axes, can also be set using `axes=dict(xygrid=True)` 820 ztolerance : (float) 821 a tolerance factor to superimpose objects (along the z-axis). 822 823 Examples: 824 - [histo_1d_a.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_1d_a.py) 825 - [histo_1d_b.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_1d_b.py) 826 - [histo_1d_c.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_1d_c.py) 827 - [histo_1d_d.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_1d_d.py) 828 829 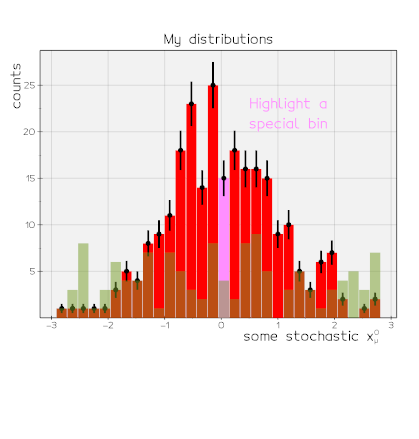 830 """ 831 832 # purge NaN from data 833 valid_ids = np.all(np.logical_not(np.isnan(data))) 834 data = np.asarray(data[valid_ids]).ravel() 835 836 # if data.dtype is integer try to center bins by default 837 if like is None and bins is None and np.issubdtype(data.dtype, np.integer): 838 if xlim is None and ylim == (0, None): 839 x1, x0 = data.max(), data.min() 840 if 0 < x1 - x0 <= 100: 841 bins = x1 - x0 + 1 842 xlim = (x0 - 0.5, x1 + 0.5) 843 844 if like is None and vedo.last_figure is not None: 845 if xlim is None and ylim == (0, None): 846 like = vedo.last_figure 847 848 if like is not None: 849 xlim = like.xlim 850 ylim = like.ylim 851 aspect = like.aspect 852 padding = like.padding 853 if bins is None: 854 bins = like.bins 855 if bins is None: 856 bins = 20 857 858 if utils.is_sequence(xlim): 859 # deal with user passing eg [x0, None] 860 _x0, _x1 = xlim 861 if _x0 is None: 862 _x0 = data.min() 863 if _x1 is None: 864 _x1 = data.max() 865 xlim = [_x0, _x1] 866 867 fs, edges = np.histogram(data, bins=bins, weights=weights, range=xlim) 868 binsize = edges[1] - edges[0] 869 ntot = data.shape[0] 870 871 fig_kwargs["title"] = title 872 fig_kwargs["xtitle"] = xtitle 873 fig_kwargs["ytitle"] = ytitle 874 fig_kwargs["ac"] = ac 875 fig_kwargs["ztolerance"] = ztolerance 876 fig_kwargs["grid"] = grid 877 878 unscaled_errors = np.sqrt(fs) 879 if density: 880 scaled_errors = unscaled_errors / (ntot * binsize) 881 fs = fs / (ntot * binsize) 882 if ytitle == " ": 883 ytitle = f"counts / ({ntot} x {utils.precision(binsize,3)})" 884 fig_kwargs["ytitle"] = ytitle 885 elif logscale: 886 se_up = np.log10(fs + unscaled_errors / 2 + 1) 887 se_dw = np.log10(fs - unscaled_errors / 2 + 1) 888 scaled_errors = np.c_[se_up, se_dw] 889 fs = np.log10(fs + 1) 890 if ytitle == " ": 891 ytitle = "log_10 (counts+1)" 892 fig_kwargs["ytitle"] = ytitle 893 894 x0, x1 = np.min(edges), np.max(edges) 895 y0, y1 = ylim[0], np.max(fs) 896 897 _errors = [] 898 if errors: 899 if density: 900 y1 += max(scaled_errors) / 2 901 _errors = scaled_errors 902 elif logscale: 903 y1 = max(scaled_errors[:, 0]) 904 _errors = scaled_errors 905 else: 906 y1 += max(unscaled_errors) / 2 907 _errors = unscaled_errors 908 909 if like is None: 910 ylim = list(ylim) 911 if xlim is None: 912 xlim = [x0, x1] 913 if ylim[1] is None: 914 ylim[1] = y1 915 if ylim[0] != 0: 916 ylim[0] = y0 917 918 self.title = title 919 self.xtitle = xtitle 920 self.ytitle = ytitle 921 self.entries = ntot 922 self.frequencies = fs 923 self.errors = _errors 924 self.edges = edges 925 self.centers = (edges[0:-1] + edges[1:]) / 2 926 self.mean = data.mean() 927 self.std = data.std() 928 self.bins = edges # internally used by "like" 929 930 ############################### stats legend as htitle 931 addstats = False 932 if not title: 933 if "axes" not in fig_kwargs: 934 addstats = True 935 axes_opts = {} 936 fig_kwargs["axes"] = axes_opts 937 elif fig_kwargs["axes"] is False: 938 pass 939 else: 940 axes_opts = fig_kwargs["axes"] 941 if "htitle" not in axes_opts: 942 addstats = True 943 944 if addstats: 945 htitle = f"Entries:~~{int(self.entries)} " 946 htitle += f"Mean:~~{utils.precision(self.mean, 4)} " 947 htitle += f"STD:~~{utils.precision(self.std, 4)} " 948 949 axes_opts["htitle"] = htitle 950 axes_opts["htitle_justify"] = "bottom-left" 951 axes_opts["htitle_size"] = 0.016 952 axes_opts["htitle_offset"] = [-0.49, 0.01, 0] 953 954 if mc is None: 955 mc = lc 956 if ma is None: 957 ma = alpha 958 959 if label: 960 nlab = LabelData() 961 nlab.text = label 962 nlab.tcolor = ac 963 nlab.marker = marker 964 nlab.mcolor = mc 965 if not marker: 966 nlab.marker = "s" 967 nlab.mcolor = c 968 fig_kwargs["label"] = nlab 969 970 ############################################### Figure init 971 Figure.__init__(self, xlim, ylim, aspect, padding, **fig_kwargs) 972 if not self.yscale: 973 return 974 975 if utils.is_sequence(bins): 976 myedges = np.array(bins) 977 bins = len(bins) - 1 978 else: 979 myedges = edges 980 981 bin_centers = [] 982 for i in range(bins): 983 x = (myedges[i] + myedges[i + 1]) / 2 984 bin_centers.append([x, fs[i], 0]) 985 986 rs = [] 987 maxheigth = 0 988 if not fill and not outline and not errors and not marker: 989 outline = True # otherwise it's empty.. 990 991 if fill: ##################### 992 if outline: 993 gap = 0 994 995 for i in range(bins): 996 F = fs[i] 997 if not F: 998 continue 999 p0 = (myedges[i] + gap * binsize, 0, 0) 1000 p1 = (myedges[i + 1] - gap * binsize, F, 0) 1001 1002 if radius: 1003 if gap: 1004 rds = np.array([0, 0, radius, radius]) 1005 else: 1006 rd1 = 0 if i < bins - 1 and fs[i + 1] >= F else radius / 2 1007 rd2 = 0 if i > 0 and fs[i - 1] >= F else radius / 2 1008 rds = np.array([0, 0, rd1, rd2]) 1009 p1_yscaled = [p1[0], p1[1] * self.yscale, 0] 1010 r = shapes.Rectangle(p0, p1_yscaled, radius=rds * binsize, res=6) 1011 r.scale([1, 1 / self.yscale, 1]) 1012 r.radius = None # so it doesnt get recreated and rescaled by insert() 1013 else: 1014 r = shapes.Rectangle(p0, p1) 1015 1016 if texture: 1017 r.texture(texture) 1018 c = "w" 1019 # if texture: # causes Segmentation fault vtk9.0.3 1020 # if i>0 and rs[i-1].GetTexture(): # reuse the same texture obj 1021 # r.texture(rs[i-1].GetTexture()) 1022 # else: 1023 # r.texture(texture) 1024 # c = 'w' 1025 1026 r.PickableOff() 1027 maxheigth = max(maxheigth, p1[1]) 1028 if c in colors.cmaps_names: 1029 col = colors.color_map((p0[0] + p1[0]) / 2, c, myedges[0], myedges[-1]) 1030 else: 1031 col = c 1032 r.color(col).alpha(alpha).lighting("off") 1033 r.z(self.ztolerance) 1034 rs.append(r) 1035 1036 if outline: ##################### 1037 lns = [[myedges[0], 0, 0]] 1038 for i in range(bins): 1039 lns.append([myedges[i], fs[i], 0]) 1040 lns.append([myedges[i + 1], fs[i], 0]) 1041 maxheigth = max(maxheigth, fs[i]) 1042 lns.append([myedges[-1], 0, 0]) 1043 outl = shapes.Line(lns, c=lc, alpha=alpha, lw=lw) 1044 outl.z(self.ztolerance * 2) 1045 rs.append(outl) 1046 1047 if errors: ##################### 1048 for i in range(bins): 1049 x = self.centers[i] 1050 f = fs[i] 1051 if not f: 1052 continue 1053 err = _errors[i] 1054 if utils.is_sequence(err): 1055 el = shapes.Line([x, err[0], 0], [x, err[1], 0], c=lc, alpha=alpha, lw=lw) 1056 else: 1057 el = shapes.Line( 1058 [x, f - err / 2, 0], [x, f + err / 2, 0], c=lc, alpha=alpha, lw=lw 1059 ) 1060 el.z(self.ztolerance * 3) 1061 rs.append(el) 1062 1063 if marker: ##################### 1064 1065 # remove empty bins (we dont want a marker there) 1066 bin_centers = np.array(bin_centers) 1067 bin_centers = bin_centers[bin_centers[:, 1] > 0] 1068 1069 if utils.is_sequence(ms): ### variable point size 1070 mk = shapes.Marker(marker, s=1) 1071 mk.scale([1, 1 / self.yscale, 1]) 1072 msv = np.zeros_like(bin_centers) 1073 msv[:, 0] = ms 1074 marked = shapes.Glyph( 1075 bin_centers, mk, c=mc, orientation_array=msv, scale_by_vector_size=True 1076 ) 1077 else: ### fixed point size 1078 1079 if ms is None: 1080 ms = (xlim[1] - xlim[0]) / 100.0 1081 else: 1082 ms = (xlim[1] - xlim[0]) / 100.0 * ms 1083 1084 if utils.is_sequence(mc): 1085 mk = shapes.Marker(marker, s=ms) 1086 mk.scale([1, 1 / self.yscale, 1]) 1087 msv = np.zeros_like(bin_centers) 1088 msv[:, 0] = 1 1089 marked = shapes.Glyph( 1090 bin_centers, mk, c=mc, orientation_array=msv, scale_by_vector_size=True 1091 ) 1092 else: 1093 mk = shapes.Marker(marker, s=ms) 1094 mk.scale([1, 1 / self.yscale, 1]) 1095 marked = shapes.Glyph(bin_centers, mk, c=mc) 1096 1097 marked.alpha(ma) 1098 marked.z(self.ztolerance * 4) 1099 rs.append(marked) 1100 1101 self.insert(*rs, as3d=False) 1102 self.name = "Histogram1D" 1103 1104 def print(self, **kwargs): 1105 """Print infos about this histogram""" 1106 txt = ( 1107 f"{self.name} {self.title}\n" 1108 f" xtitle = '{self.xtitle}'\n" 1109 f" ytitle = '{self.ytitle}'\n" 1110 f" entries = {self.entries}\n" 1111 f" mean = {self.mean}\n" 1112 f" std = {self.std}" 1113 ) 1114 colors.printc(txt, **kwargs) 1115 1116 1117######################################################################################### 1118class Histogram2D(Figure): 1119 """2D histogramming.""" 1120 1121 def __init__( 1122 self, 1123 xvalues, 1124 yvalues=None, 1125 bins=25, 1126 weights=None, 1127 cmap="cividis", 1128 alpha=1, 1129 gap=0, 1130 scalarbar=True, 1131 # Figure and axes options: 1132 like=None, 1133 xlim=None, 1134 ylim=(None, None), 1135 zlim=(None, None), 1136 aspect=1, 1137 title="", 1138 xtitle=" ", 1139 ytitle=" ", 1140 ztitle="", 1141 ac="k", 1142 **fig_kwargs, 1143 ): 1144 """ 1145 Input data formats `[(x1,x2,..), (y1,y2,..)] or [(x1,y1), (x2,y2),..]` 1146 are both valid. 1147 1148 Use keyword `like=...` if you want to use the same format of a previously 1149 created Figure (useful when superimposing Figures) to make sure 1150 they are compatible and comparable. If they are not compatible 1151 you will receive an error message. 1152 1153 Arguments: 1154 bins : (list) 1155 binning as (nx, ny) 1156 weights : (list) 1157 array of weights to assign to each entry 1158 cmap : (str, lookuptable) 1159 color map name or look up table 1160 alpha : (float) 1161 opacity of the histogram 1162 gap : (float) 1163 separation between adjacent bins as a fraction for their size 1164 scalarbar : (bool) 1165 add a scalarbar to right of the histogram 1166 like : (Figure) 1167 grab and use the same format of the given Figure (for superimposing) 1168 xlim : (list) 1169 [x0, x1] range of interest. If left to None will automatically 1170 choose the minimum or the maximum of the data range. 1171 Data outside the range are completely ignored. 1172 ylim : (list) 1173 [y0, y1] range of interest. If left to None will automatically 1174 choose the minimum or the maximum of the data range. 1175 Data outside the range are completely ignored. 1176 aspect : (float) 1177 the desired aspect ratio of the figure. 1178 title : (str) 1179 title of the plot to appear on top. 1180 If left blank some statistics will be shown. 1181 xtitle : (str) 1182 x axis title 1183 ytitle : (str) 1184 y axis title 1185 ztitle : (str) 1186 title for the scalar bar 1187 ac : (str) 1188 axes color, additional keyword for Axes can also be added 1189 using e.g. `axes=dict(xygrid=True)` 1190 1191 Examples: 1192 - [histo_2d_a.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_2d_a.py) 1193 - [histo_2d_b.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_2d_b.py) 1194 1195 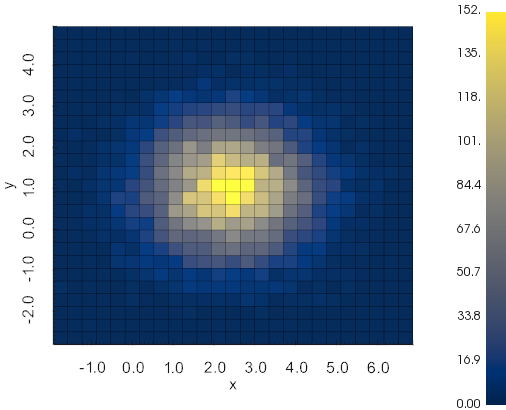 1196 """ 1197 xvalues = np.asarray(xvalues) 1198 if yvalues is None: 1199 # assume [(x1,y1), (x2,y2) ...] format 1200 yvalues = xvalues[:, 1] 1201 xvalues = xvalues[:, 0] 1202 else: 1203 yvalues = np.asarray(yvalues) 1204 1205 padding = [0, 0, 0, 0] 1206 1207 if like is None and vedo.last_figure is not None: 1208 if xlim is None and ylim == (None, None) and zlim == (None, None): 1209 like = vedo.last_figure 1210 1211 if like is not None: 1212 xlim = like.xlim 1213 ylim = like.ylim 1214 aspect = like.aspect 1215 padding = like.padding 1216 if bins is None: 1217 bins = like.bins 1218 if bins is None: 1219 bins = 20 1220 1221 if isinstance(bins, int): 1222 bins = (bins, bins) 1223 1224 if utils.is_sequence(xlim): 1225 # deal with user passing eg [x0, None] 1226 _x0, _x1 = xlim 1227 if _x0 is None: 1228 _x0 = xvalues.min() 1229 if _x1 is None: 1230 _x1 = xvalues.max() 1231 xlim = [_x0, _x1] 1232 1233 if utils.is_sequence(ylim): 1234 # deal with user passing eg [x0, None] 1235 _y0, _y1 = ylim 1236 if _y0 is None: 1237 _y0 = yvalues.min() 1238 if _y1 is None: 1239 _y1 = yvalues.max() 1240 ylim = [_y0, _y1] 1241 1242 H, xedges, yedges = np.histogram2d( 1243 xvalues, yvalues, weights=weights, bins=bins, range=(xlim, ylim) 1244 ) 1245 1246 xlim = np.min(xedges), np.max(xedges) 1247 ylim = np.min(yedges), np.max(yedges) 1248 dx, dy = xlim[1] - xlim[0], ylim[1] - ylim[0] 1249 1250 fig_kwargs["title"] = title 1251 fig_kwargs["xtitle"] = xtitle 1252 fig_kwargs["ytitle"] = ytitle 1253 fig_kwargs["ac"] = ac 1254 1255 self.entries = len(xvalues) 1256 self.frequencies = H 1257 self.edges = (xedges, yedges) 1258 self.mean = (xvalues.mean(), yvalues.mean()) 1259 self.std = (xvalues.std(), yvalues.std()) 1260 self.bins = bins # internally used by "like" 1261 1262 ############################### stats legend as htitle 1263 addstats = False 1264 if not title: 1265 if "axes" not in fig_kwargs: 1266 addstats = True 1267 axes_opts = {} 1268 fig_kwargs["axes"] = axes_opts 1269 elif fig_kwargs["axes"] is False: 1270 pass 1271 else: 1272 axes_opts = fig_kwargs["axes"] 1273 if "htitle" not in fig_kwargs["axes"]: 1274 addstats = True 1275 1276 if addstats: 1277 htitle = f"Entries:~~{int(self.entries)} " 1278 htitle += f"Mean:~~{utils.precision(self.mean, 3)} " 1279 htitle += f"STD:~~{utils.precision(self.std, 3)} " 1280 axes_opts["htitle"] = htitle 1281 axes_opts["htitle_justify"] = "bottom-left" 1282 axes_opts["htitle_size"] = 0.0175 1283 axes_opts["htitle_offset"] = [-0.49, 0.01, 0] 1284 1285 ############################################### Figure init 1286 Figure.__init__(self, xlim, ylim, aspect, padding, **fig_kwargs) 1287 1288 if self.yscale: 1289 ##################### the grid 1290 acts = [] 1291 g = shapes.Grid( 1292 pos=[(xlim[0] + xlim[1]) / 2, (ylim[0] + ylim[1]) / 2, 0], s=(dx, dy), res=bins[:2] 1293 ) 1294 g.alpha(alpha).lw(0).wireframe(False).flat().lighting("off") 1295 g.cmap(cmap, np.ravel(H.T), on="cells", vmin=zlim[0], vmax=zlim[1]) 1296 if gap: 1297 g.shrink(abs(1 - gap)) 1298 1299 if scalarbar: 1300 sc = g.add_scalarbar3d(ztitle, c=ac).scalarbar 1301 sc.scale([self.yscale, 1, 1]) ## prescale trick 1302 sbnds = sc.xbounds() 1303 sc.x(self.x1lim + (sbnds[1] - sbnds[0]) * 0.75) 1304 acts.append(sc) 1305 acts.append(g) 1306 1307 self.insert(*acts, as3d=False) 1308 self.name = "Histogram2D" 1309 1310 1311######################################################################################### 1312class PlotBars(Figure): 1313 """Creates a `PlotBars(Figure)` object.""" 1314 1315 def __init__( 1316 self, 1317 data, 1318 errors=False, 1319 logscale=False, 1320 fill=True, 1321 gap=0.02, 1322 radius=0.05, 1323 c="olivedrab", 1324 alpha=1, 1325 texture="", 1326 outline=False, 1327 lw=2, 1328 lc="k", 1329 # Figure and axes options: 1330 like=None, 1331 xlim=(None, None), 1332 ylim=(0, None), 1333 aspect=4 / 3, 1334 padding=(0.025, 0.025, 0, 0.05), 1335 # 1336 title="", 1337 xtitle=" ", 1338 ytitle=" ", 1339 ac="k", 1340 grid=False, 1341 ztolerance=None, 1342 **fig_kwargs, 1343 ): 1344 """ 1345 Input must be in format `[counts, labels, colors, edges]`. 1346 Either or both `edges` and `colors` are optional and can be omitted. 1347 1348 Use keyword `like=...` if you want to use the same format of a previously 1349 created Figure (useful when superimposing Figures) to make sure 1350 they are compatible and comparable. If they are not compatible 1351 you will receive an error message. 1352 1353 Arguments: 1354 errors : (bool) 1355 show error bars 1356 logscale : (bool) 1357 use logscale on y-axis 1358 fill : (bool) 1359 fill bars with solid color `c` 1360 gap : (float) 1361 leave a small space btw bars 1362 radius : (float) 1363 border radius of the top of the histogram bar. Default value is 0.1. 1364 texture : (str) 1365 url or path to an image to be used as texture for the bin 1366 outline : (bool) 1367 show outline of the bins 1368 xtitle : (str) 1369 title for the x-axis, can also be set using `axes=dict(xtitle="my x axis")` 1370 ytitle : (str) 1371 title for the y-axis, can also be set using `axes=dict(ytitle="my y axis")` 1372 ac : (str) 1373 axes color 1374 padding : (float, list) 1375 keep a padding space from the axes (as a fraction of the axis size). 1376 This can be a list of four numbers. 1377 aspect : (float) 1378 the desired aspect ratio of the figure. Default is 4/3. 1379 grid : (bool) 1380 show the background grid for the axes, can also be set using `axes=dict(xygrid=True)` 1381 1382 Examples: 1383 - [plot_bars.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/plot_bars.py) 1384 1385 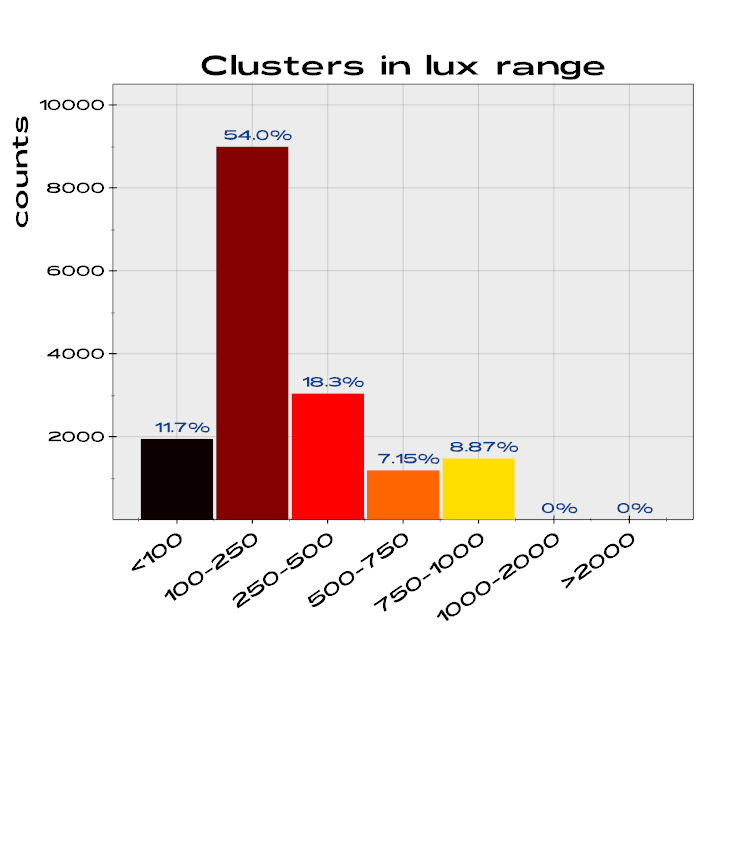 1386 """ 1387 ndata = len(data) 1388 if ndata == 4: 1389 counts, xlabs, cols, edges = data 1390 elif ndata == 3: 1391 counts, xlabs, cols = data 1392 edges = np.array(range(len(counts) + 1)) + 0.5 1393 elif ndata == 2: 1394 counts, xlabs = data 1395 edges = np.array(range(len(counts) + 1)) + 0.5 1396 cols = [c] * len(counts) 1397 else: 1398 m = "barplot error: data must be given as [counts, labels, colors, edges] not\n" 1399 vedo.logger.error(m + f" {data}\n bin edges and colors are optional.") 1400 raise RuntimeError() 1401 1402 # sanity checks 1403 assert len(counts) == len(xlabs) 1404 assert len(counts) == len(cols) 1405 assert len(counts) == len(edges) - 1 1406 1407 counts = np.asarray(counts) 1408 edges = np.asarray(edges) 1409 1410 if logscale: 1411 counts = np.log10(counts + 1) 1412 if ytitle == " ": 1413 ytitle = "log_10 (counts+1)" 1414 1415 if like is None and vedo.last_figure is not None: 1416 if xlim == (None, None) and ylim == (0, None): 1417 like = vedo.last_figure 1418 1419 if like is not None: 1420 xlim = like.xlim 1421 ylim = like.ylim 1422 aspect = like.aspect 1423 padding = like.padding 1424 1425 if utils.is_sequence(xlim): 1426 # deal with user passing eg [x0, None] 1427 _x0, _x1 = xlim 1428 if _x0 is None: 1429 _x0 = np.min(edges) 1430 if _x1 is None: 1431 _x1 = np.max(edges) 1432 xlim = [_x0, _x1] 1433 1434 x0, x1 = np.min(edges), np.max(edges) 1435 y0, y1 = ylim[0], np.max(counts) 1436 1437 if like is None: 1438 ylim = list(ylim) 1439 if xlim is None: 1440 xlim = [x0, x1] 1441 if ylim[1] is None: 1442 ylim[1] = y1 1443 if ylim[0] != 0: 1444 ylim[0] = y0 1445 1446 fig_kwargs["title"] = title 1447 fig_kwargs["xtitle"] = xtitle 1448 fig_kwargs["ytitle"] = ytitle 1449 fig_kwargs["ac"] = ac 1450 fig_kwargs["ztolerance"] = ztolerance 1451 fig_kwargs["grid"] = grid 1452 1453 centers = (edges[0:-1] + edges[1:]) / 2 1454 binsizes = (centers - edges[0:-1]) * 2 1455 1456 if "axes" not in fig_kwargs: 1457 fig_kwargs["axes"] = {} 1458 1459 _xlabs = [] 1460 for center, xlb in zip(centers, xlabs): 1461 _xlabs.append([center, str(xlb)]) 1462 fig_kwargs["axes"]["x_values_and_labels"] = _xlabs 1463 1464 ############################################### Figure 1465 self.statslegend = "" 1466 self.edges = edges 1467 self.centers = centers 1468 self.bins = edges # internal used by "like" 1469 Figure.__init__(self, xlim, ylim, aspect, padding, **fig_kwargs) 1470 if not self.yscale: 1471 return 1472 1473 rs = [] 1474 maxheigth = 0 1475 if fill: ##################### 1476 if outline: 1477 gap = 0 1478 1479 for i in range(len(centers)): 1480 binsize = binsizes[i] 1481 p0 = (edges[i] + gap * binsize, 0, 0) 1482 p1 = (edges[i + 1] - gap * binsize, counts[i], 0) 1483 1484 if radius: 1485 rds = np.array([0, 0, radius, radius]) 1486 p1_yscaled = [p1[0], p1[1] * self.yscale, 0] 1487 r = shapes.Rectangle(p0, p1_yscaled, radius=rds * binsize, res=6) 1488 r.scale([1, 1 / self.yscale, 1]) 1489 r.radius = None # so it doesnt get recreated and rescaled by insert() 1490 else: 1491 r = shapes.Rectangle(p0, p1) 1492 1493 if texture: 1494 r.texture(texture) 1495 c = "w" 1496 1497 r.PickableOff() 1498 maxheigth = max(maxheigth, p1[1]) 1499 if c in colors.cmaps_names: 1500 col = colors.color_map((p0[0] + p1[0]) / 2, c, edges[0], edges[-1]) 1501 else: 1502 col = cols[i] 1503 r.color(col).alpha(alpha).lighting("off") 1504 r.name = f"bar_{i}" 1505 r.z(self.ztolerance) 1506 rs.append(r) 1507 1508 elif outline: ##################### 1509 lns = [[edges[0], 0, 0]] 1510 for i in range(len(centers)): 1511 lns.append([edges[i], counts[i], 0]) 1512 lns.append([edges[i + 1], counts[i], 0]) 1513 maxheigth = max(maxheigth, counts[i]) 1514 lns.append([edges[-1], 0, 0]) 1515 outl = shapes.Line(lns, c=lc, alpha=alpha, lw=lw).z(self.ztolerance) 1516 outl.name = f"bar_outline_{i}" 1517 rs.append(outl) 1518 1519 if errors: ##################### 1520 for x, f in centers: 1521 err = np.sqrt(f) 1522 el = shapes.Line([x, f - err / 2, 0], [x, f + err / 2, 0], c=lc, alpha=alpha, lw=lw) 1523 el.z(self.ztolerance * 2) 1524 rs.append(el) 1525 1526 self.insert(*rs, as3d=False) 1527 self.name = "PlotBars" 1528 1529 1530######################################################################################### 1531class PlotXY(Figure): 1532 """Creates a `PlotXY(Figure)` object.""" 1533 1534 def __init__( 1535 self, 1536 # 1537 data, 1538 xerrors=None, 1539 yerrors=None, 1540 # 1541 lw=2, 1542 lc=None, 1543 la=1, 1544 dashed=False, 1545 splined=False, 1546 # 1547 elw=2, # error line width 1548 ec=None, # error line or band color 1549 error_band=False, # errors in x are ignored 1550 # 1551 marker="", 1552 ms=None, 1553 mc=None, 1554 ma=None, 1555 # Figure and axes options: 1556 like=None, 1557 xlim=None, 1558 ylim=(None, None), 1559 aspect=4 / 3, 1560 padding=0.05, 1561 # 1562 title="", 1563 xtitle=" ", 1564 ytitle=" ", 1565 ac="k", 1566 grid=True, 1567 ztolerance=None, 1568 label="", 1569 **fig_kwargs, 1570 ): 1571 """ 1572 Arguments: 1573 xerrors : (bool) 1574 show error bars associated to each point in x 1575 yerrors : (bool) 1576 show error bars associated to each point in y 1577 lw : (int) 1578 width of the line connecting points in pixel units. 1579 Set it to 0 to remove the line. 1580 lc : (str) 1581 line color 1582 la : (float) 1583 line "alpha", opacity of the line 1584 dashed : (bool) 1585 draw a dashed line instead of a continuous line 1586 splined : (bool) 1587 spline the line joining the point as a countinous curve 1588 elw : (int) 1589 width of error bar lines in units of pixels 1590 ec : (color) 1591 color of error bar, by default the same as marker color 1592 error_band : (bool) 1593 represent errors on y as a filled error band. 1594 Use `ec` keyword to modify its color. 1595 marker : (str, int) 1596 use a marker for the data points 1597 ms : (float) 1598 marker size 1599 mc : (color) 1600 color of the marker 1601 ma : (float) 1602 opacity of the marker 1603 xlim : (list) 1604 set limits to the range for the x variable 1605 ylim : (list) 1606 set limits to the range for the y variable 1607 aspect : (float, str) 1608 Desired aspect ratio. 1609 Use `aspect="equal"` to force the same units in x and y. 1610 Scaling factor is saved in Figure.yscale. 1611 padding : (float, list) 1612 keep a padding space from the axes (as a fraction of the axis size). 1613 This can be a list of four numbers. 1614 title : (str) 1615 title to appear on the top of the frame, like a header. 1616 xtitle : (str) 1617 title for the x-axis, can also be set using `axes=dict(xtitle="my x axis")` 1618 ytitle : (str) 1619 title for the y-axis, can also be set using `axes=dict(ytitle="my y axis")` 1620 ac : (str) 1621 axes color 1622 grid : (bool) 1623 show the background grid for the axes, can also be set using `axes=dict(xygrid=True)` 1624 ztolerance : (float) 1625 a tolerance factor to superimpose objects (along the z-axis). 1626 1627 Example: 1628 ```python 1629 import numpy as np 1630 from vedo.pyplot import plot 1631 x = np.arange(0, np.pi, 0.1) 1632 fig = plot(x, np.sin(2*x), 'r0-', aspect='equal') 1633 fig+= plot(x, np.cos(2*x), 'blue4 o-', like=fig) 1634 fig.show().close() 1635 ``` 1636 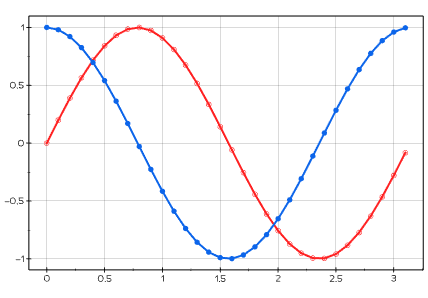 1637 1638 Examples: 1639 - [plot_errbars.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/plot_errbars.py) 1640 - [plot_errband.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/plot_errband.py) 1641 - [plot_pip.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/plot_pip.py) 1642 1643 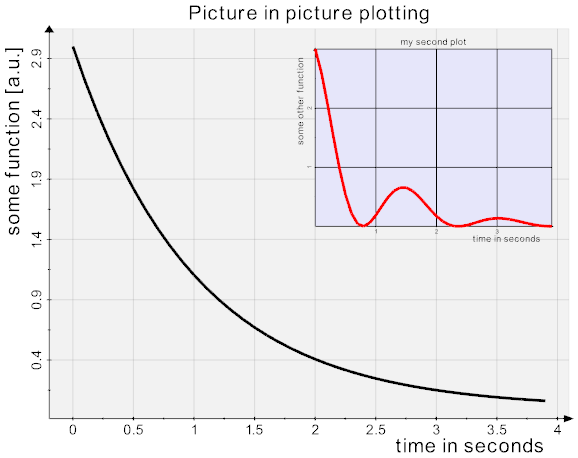 1644 1645 - [scatter1.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/scatter1.py) 1646 - [scatter2.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/scatter2.py) 1647 1648 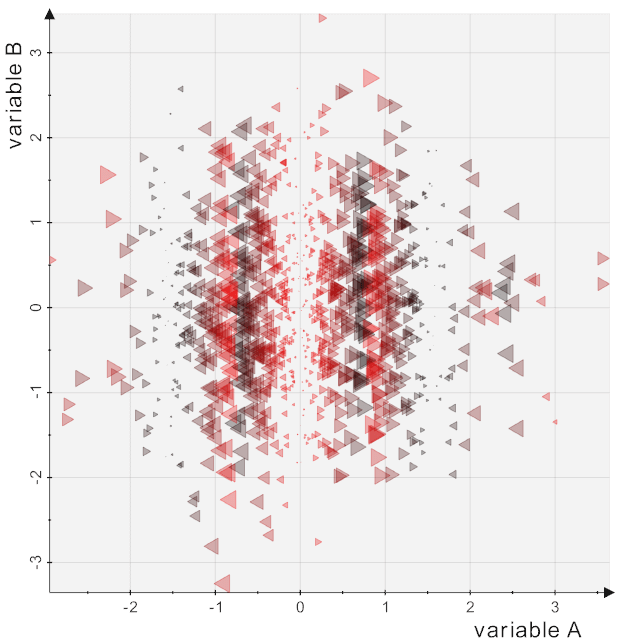 1649 """ 1650 line = False 1651 if lw > 0: 1652 line = True 1653 if marker == "" and not line and not splined: 1654 marker = "o" 1655 1656 if like is None and vedo.last_figure is not None: 1657 if xlim is None and ylim == (None, None): 1658 like = vedo.last_figure 1659 1660 if like is not None: 1661 xlim = like.xlim 1662 ylim = like.ylim 1663 aspect = like.aspect 1664 padding = like.padding 1665 1666 if utils.is_sequence(xlim): 1667 # deal with user passing eg [x0, None] 1668 _x0, _x1 = xlim 1669 if _x0 is None: 1670 _x0 = data.min() 1671 if _x1 is None: 1672 _x1 = data.max() 1673 xlim = [_x0, _x1] 1674 1675 # purge NaN from data 1676 validIds = np.all(np.logical_not(np.isnan(data))) 1677 data = np.array(data[validIds])[0] 1678 1679 fig_kwargs["title"] = title 1680 fig_kwargs["xtitle"] = xtitle 1681 fig_kwargs["ytitle"] = ytitle 1682 fig_kwargs["ac"] = ac 1683 fig_kwargs["ztolerance"] = ztolerance 1684 fig_kwargs["grid"] = grid 1685 1686 x0, y0 = np.min(data, axis=0) 1687 x1, y1 = np.max(data, axis=0) 1688 if xerrors is not None and not error_band: 1689 x0 = min(data[:, 0] - xerrors) 1690 x1 = max(data[:, 0] + xerrors) 1691 if yerrors is not None: 1692 y0 = min(data[:, 1] - yerrors) 1693 y1 = max(data[:, 1] + yerrors) 1694 1695 if like is None: 1696 if xlim is None: 1697 xlim = (None, None) 1698 xlim = list(xlim) 1699 if xlim[0] is None: 1700 xlim[0] = x0 1701 if xlim[1] is None: 1702 xlim[1] = x1 1703 ylim = list(ylim) 1704 if ylim[0] is None: 1705 ylim[0] = y0 1706 if ylim[1] is None: 1707 ylim[1] = y1 1708 1709 self.entries = len(data) 1710 self.mean = data.mean() 1711 self.std = data.std() 1712 1713 ######### the PlotXY marker 1714 # fall back solutions logic for colors 1715 if "c" in fig_kwargs: 1716 if mc is None: 1717 mc = fig_kwargs["c"] 1718 if lc is None: 1719 lc = fig_kwargs["c"] 1720 if ec is None: 1721 ec = fig_kwargs["c"] 1722 if lc is None: 1723 lc = "k" 1724 if mc is None: 1725 mc = lc 1726 if ma is None: 1727 ma = la 1728 if ec is None: 1729 if mc is None: 1730 ec = lc 1731 else: 1732 ec = mc 1733 1734 if label: 1735 nlab = LabelData() 1736 nlab.text = label 1737 nlab.tcolor = ac 1738 nlab.marker = marker 1739 if line and marker == "": 1740 nlab.marker = "-" 1741 nlab.mcolor = mc 1742 fig_kwargs["label"] = nlab 1743 1744 ############################################### Figure init 1745 Figure.__init__(self, xlim, ylim, aspect, padding, **fig_kwargs) 1746 1747 if not self.yscale: 1748 return 1749 1750 acts = [] 1751 1752 ######### the PlotXY Line or Spline 1753 if dashed: 1754 l = shapes.DashedLine(data, c=lc, alpha=la, lw=lw) 1755 acts.append(l) 1756 elif splined: 1757 l = shapes.KSpline(data).lw(lw).c(lc).alpha(la) 1758 acts.append(l) 1759 elif line: 1760 l = shapes.Line(data, c=lc, alpha=la).lw(lw) 1761 acts.append(l) 1762 1763 if marker: 1764 1765 pts = np.c_[data, np.zeros(len(data))] 1766 1767 if utils.is_sequence(ms): 1768 ### variable point size 1769 mk = shapes.Marker(marker, s=1) 1770 mk.scale([1, 1 / self.yscale, 1]) 1771 msv = np.zeros_like(pts) 1772 msv[:, 0] = ms 1773 marked = shapes.Glyph( 1774 pts, mk, c=mc, orientation_array=msv, scale_by_vector_size=True 1775 ) 1776 else: 1777 ### fixed point size 1778 if ms is None: 1779 ms = (xlim[1] - xlim[0]) / 100.0 1780 1781 if utils.is_sequence(mc): 1782 fig_kwargs["marker_color"] = None # for labels 1783 mk = shapes.Marker(marker, s=ms) 1784 mk.scale([1, 1 / self.yscale, 1]) 1785 msv = np.zeros_like(pts) 1786 msv[:, 0] = 1 1787 marked = shapes.Glyph( 1788 pts, mk, c=mc, orientation_array=msv, scale_by_vector_size=True 1789 ) 1790 else: 1791 mk = shapes.Marker(marker, s=ms) 1792 mk.scale([1, 1 / self.yscale, 1]) 1793 marked = shapes.Glyph(pts, mk, c=mc) 1794 1795 marked.name = "Marker" 1796 marked.alpha(ma) 1797 marked.z(3 * self.ztolerance) 1798 acts.append(marked) 1799 1800 ######### the PlotXY marker errors 1801 ztol = self.ztolerance 1802 1803 if error_band: 1804 yerrors = np.abs(yerrors) 1805 du = np.array(data) 1806 dd = np.array(data) 1807 du[:, 1] += yerrors 1808 dd[:, 1] -= yerrors 1809 if splined: 1810 res = len(data) * 20 1811 band1 = shapes.KSpline(du, res=res) 1812 band2 = shapes.KSpline(dd, res=res) 1813 band = shapes.Ribbon(band1, band2, res=(res, 2)) 1814 else: 1815 dd = list(reversed(dd.tolist())) 1816 band = shapes.Line(du.tolist() + dd, closed=True) 1817 band.triangulate().lw(0) 1818 if ec is None: 1819 band.c(lc) 1820 else: 1821 band.c(ec) 1822 band.lighting("off").alpha(la).z(ztol / 20) 1823 acts.append(band) 1824 1825 else: 1826 1827 ## xerrors 1828 if xerrors is not None: 1829 if len(xerrors) == len(data): 1830 errs = [] 1831 for i, val in enumerate(data): 1832 xval, yval = val 1833 xerr = xerrors[i] / 2 1834 el = shapes.Line((xval - xerr, yval, ztol), (xval + xerr, yval, ztol)) 1835 el.lw(elw) 1836 errs.append(el) 1837 mxerrs = merge(errs).c(ec).lw(lw).alpha(ma).z(2 * ztol) 1838 acts.append(mxerrs) 1839 else: 1840 vedo.logger.error("in PlotXY(xerrors=...): mismatch in array length") 1841 1842 ## yerrors 1843 if yerrors is not None: 1844 if len(yerrors) == len(data): 1845 errs = [] 1846 for i, val in enumerate(data): 1847 xval, yval = val 1848 yerr = yerrors[i] 1849 el = shapes.Line((xval, yval - yerr, ztol), (xval, yval + yerr, ztol)) 1850 el.lw(elw) 1851 errs.append(el) 1852 myerrs = merge(errs).c(ec).lw(lw).alpha(ma).z(2 * ztol) 1853 acts.append(myerrs) 1854 else: 1855 vedo.logger.error("in PlotXY(yerrors=...): mismatch in array length") 1856 1857 self.insert(*acts, as3d=False) 1858 self.name = "PlotXY" 1859 1860 1861def plot(*args, **kwargs): 1862 """ 1863 Draw a 2D line plot, or scatter plot, of variable x vs variable y. 1864 Input format can be either `[allx], [allx, ally] or [(x1,y1), (x2,y2), ...]` 1865 1866 Use `like=...` if you want to use the same format of a previously 1867 created Figure (useful when superimposing Figures) to make sure 1868 they are compatible and comparable. If they are not compatible 1869 you will receive an error message. 1870 1871 Arguments: 1872 xerrors : (bool) 1873 show error bars associated to each point in x 1874 yerrors : (bool) 1875 show error bars associated to each point in y 1876 lw : (int) 1877 width of the line connecting points in pixel units. 1878 Set it to 0 to remove the line. 1879 lc : (str) 1880 line color 1881 la : (float) 1882 line "alpha", opacity of the line 1883 dashed : (bool) 1884 draw a dashed line instead of a continuous line 1885 splined : (bool) 1886 spline the line joining the point as a countinous curve 1887 elw : (int) 1888 width of error bar lines in units of pixels 1889 ec : (color) 1890 color of error bar, by default the same as marker color 1891 error_band : (bool) 1892 represent errors on y as a filled error band. 1893 Use `ec` keyword to modify its color. 1894 marker : (str, int) 1895 use a marker for the data points 1896 ms : (float) 1897 marker size 1898 mc : (color) 1899 color of the marker 1900 ma : (float) 1901 opacity of the marker 1902 xlim : (list) 1903 set limits to the range for the x variable 1904 ylim : (list) 1905 set limits to the range for the y variable 1906 aspect : (float) 1907 Desired aspect ratio. 1908 If None, it is automatically calculated to get a reasonable aspect ratio. 1909 Scaling factor is saved in Figure.yscale 1910 padding : (float, list) 1911 keep a padding space from the axes (as a fraction of the axis size). 1912 This can be a list of four numbers. 1913 title : (str) 1914 title to appear on the top of the frame, like a header. 1915 xtitle : (str) 1916 title for the x-axis, can also be set using `axes=dict(xtitle="my x axis")` 1917 ytitle : (str) 1918 title for the y-axis, can also be set using `axes=dict(ytitle="my y axis")` 1919 ac : (str) 1920 axes color 1921 grid : (bool) 1922 show the background grid for the axes, can also be set using `axes=dict(xygrid=True)` 1923 ztolerance : (float) 1924 a tolerance factor to superimpose objects (along the z-axis). 1925 1926 Example: 1927 ```python 1928 from vedo.pyplot import plot 1929 import numpy as np 1930 x = np.linspace(0, 6.28, num=50) 1931 plot(np.sin(x), 'r').plot(np.cos(x), 'bo-').show().close() 1932 ``` 1933 <img src="https://user-images.githubusercontent.com/32848391/74363882-c3638300-4dcb-11ea-8a78-eb492ad9711f.png" width="600"> 1934 1935 Examples: 1936 - [plot_errbars.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/plot_errbars.py) 1937 - [plot_errband.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/plot_errband.py) 1938 - [plot_pip.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/plot_pip.py) 1939 1940  1941 1942 - [scatter1.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/scatter1.py) 1943 - [scatter2.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/scatter2.py) 1944 1945 1946 1947 ------------------------------------------------------------------------- 1948 .. note:: mode="bar" 1949 1950 Creates a `PlotBars(Figure)` object. 1951 1952 Input must be in format `[counts, labels, colors, edges]`. 1953 Either or both `edges` and `colors` are optional and can be omitted. 1954 1955 Arguments: 1956 errors : (bool) 1957 show error bars 1958 logscale : (bool) 1959 use logscale on y-axis 1960 fill : (bool) 1961 fill bars with solid color `c` 1962 gap : (float) 1963 leave a small space btw bars 1964 radius : (float) 1965 border radius of the top of the histogram bar. Default value is 0.1. 1966 texture : (str) 1967 url or path to an image to be used as texture for the bin 1968 outline : (bool) 1969 show outline of the bins 1970 xtitle : (str) 1971 title for the x-axis, can also be set using `axes=dict(xtitle="my x axis")` 1972 ytitle : (str) 1973 title for the y-axis, can also be set using `axes=dict(ytitle="my y axis")` 1974 ac : (str) 1975 axes color 1976 padding : (float, list) 1977 keep a padding space from the axes (as a fraction of the axis size). 1978 This can be a list of four numbers. 1979 aspect : (float) 1980 the desired aspect ratio of the figure. Default is 4/3. 1981 grid : (bool) 1982 show the background grid for the axes, can also be set using `axes=dict(xygrid=True)` 1983 1984 Examples: 1985 - [histo_1d_a.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_1d_a.py) 1986 - [histo_1d_b.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_1d_b.py) 1987 - [histo_1d_c.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_1d_c.py) 1988 - [histo_1d_d.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_1d_d.py) 1989 1990  1991 1992 1993 ---------------------------------------------------------------------- 1994 .. note:: 2D functions 1995 1996 If input is an external function or a formula, draw the surface 1997 representing the function `f(x,y)`. 1998 1999 Arguments: 2000 x : (float) 2001 x range of values 2002 y : (float) 2003 y range of values 2004 zlimits : (float) 2005 limit the z range of the independent variable 2006 zlevels : (int) 2007 will draw the specified number of z-levels contour lines 2008 show_nan : (bool) 2009 show where the function does not exist as red points 2010 bins : (list) 2011 number of bins in x and y 2012 2013 Examples: 2014 - [plot_fxy.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/plot_fxy.py) 2015 2016 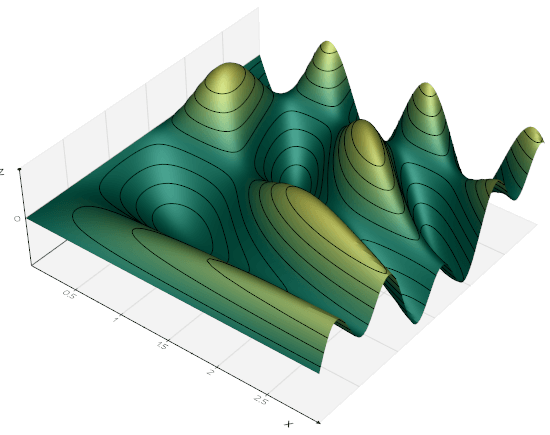 2017 2018 2019 -------------------------------------------------------------------- 2020 .. note:: mode="complex" 2021 2022 If `mode='complex'` draw the real value of the function and color map the imaginary part. 2023 2024 Arguments: 2025 cmap : (str) 2026 diverging color map (white means `imag(z)=0`) 2027 lw : (float) 2028 line with of the binning 2029 bins : (list) 2030 binning in x and y 2031 2032 Examples: 2033 - [plot_fxy.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/plot_fxy.py) 2034 2035  2036 2037 2038 -------------------------------------------------------------------- 2039 .. note:: mode="polar" 2040 2041 If `mode='polar'` input arrays are interpreted as a list of polar angles and radii. 2042 Build a polar (radar) plot by joining the set of points in polar coordinates. 2043 2044 Arguments: 2045 title : (str) 2046 plot title 2047 tsize : (float) 2048 title size 2049 bins : (int) 2050 number of bins in phi 2051 r1 : (float) 2052 inner radius 2053 r2 : (float) 2054 outer radius 2055 lsize : (float) 2056 label size 2057 c : (color) 2058 color of the line 2059 ac : (color) 2060 color of the frame and labels 2061 alpha : (float) 2062 opacity of the frame 2063 ps : (int) 2064 point size in pixels, if ps=0 no point is drawn 2065 lw : (int) 2066 line width in pixels, if lw=0 no line is drawn 2067 deg : (bool) 2068 input array is in degrees 2069 vmax : (float) 2070 normalize radius to this maximum value 2071 fill : (bool) 2072 fill convex area with solid color 2073 splined : (bool) 2074 interpolate the set of input points 2075 show_disc : (bool) 2076 draw the outer ring axis 2077 nrays : (int) 2078 draw this number of axis rays (continuous and dashed) 2079 show_lines : (bool) 2080 draw lines to the origin 2081 show_angles : (bool) 2082 draw angle values 2083 2084 Examples: 2085 - [histo_polar.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_polar.py) 2086 2087  2088 2089 2090 -------------------------------------------------------------------- 2091 .. note:: mode="spheric" 2092 2093 If `mode='spheric'` input must be an external function rho(theta, phi). 2094 A surface is created in spherical coordinates. 2095 2096 Return an `Figure(Assembly)` of 2 objects: the unit 2097 sphere (in wireframe representation) and the surface `rho(theta, phi)`. 2098 2099 Arguments: 2100 rfunc : function 2101 handle to a user defined function `rho(theta, phi)`. 2102 normalize : (bool) 2103 scale surface to fit inside the unit sphere 2104 res : (int) 2105 grid resolution of the unit sphere 2106 scalarbar : (bool) 2107 add a 3D scalarbar to the plot for radius 2108 c : (color) 2109 color of the unit sphere 2110 alpha : (float) 2111 opacity of the unit sphere 2112 cmap : (str) 2113 color map for the surface 2114 2115 Examples: 2116 - [plot_spheric.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/plot_spheric.py) 2117 2118  2119 """ 2120 mode = kwargs.pop("mode", "") 2121 if "spher" in mode: 2122 return _plot_spheric(args[0], **kwargs) 2123 2124 if "bar" in mode: 2125 return PlotBars(args[0], **kwargs) 2126 2127 if isinstance(args[0], str) or "function" in str(type(args[0])): 2128 if "complex" in mode: 2129 return _plot_fz(args[0], **kwargs) 2130 return _plot_fxy(args[0], **kwargs) 2131 2132 # grab the matplotlib-like options 2133 optidx = None 2134 for i, a in enumerate(args): 2135 if i > 0 and isinstance(a, str): 2136 optidx = i 2137 break 2138 if optidx: 2139 opts = args[optidx].replace(" ", "") 2140 if "--" in opts: 2141 opts = opts.replace("--", "") 2142 kwargs["dashed"] = True 2143 elif "-" in opts: 2144 opts = opts.replace("-", "") 2145 else: 2146 kwargs["lw"] = 0 2147 2148 symbs = [".", "o", "O", "0", "p", "*", "h", "D", "d", "v", "^", ">", "<", "s", "x", "a"] 2149 2150 allcols = list(colors.colors.keys()) + list(colors.color_nicks.keys()) 2151 for cc in allcols: 2152 if cc == "o": 2153 continue 2154 if cc in opts: 2155 opts = opts.replace(cc, "") 2156 kwargs["lc"] = cc 2157 kwargs["mc"] = cc 2158 break 2159 2160 for ss in symbs: 2161 if ss in opts: 2162 opts = opts.replace(ss, "", 1) 2163 kwargs["marker"] = ss 2164 break 2165 2166 opts.replace(" ", "") 2167 if opts: 2168 vedo.logger.error(f"in plot(), could not understand option(s): {opts}") 2169 2170 if optidx == 1 or optidx is None: 2171 if utils.is_sequence(args[0][0]) and len(args[0][0]) > 1: 2172 # print('------------- case 1', 'plot([(x,y),..])') 2173 data = np.asarray(args[0]) # (x,y) 2174 x = np.asarray(data[:, 0]) 2175 y = np.asarray(data[:, 1]) 2176 2177 elif len(args) == 1 or optidx == 1: 2178 # print('------------- case 2', 'plot(x)') 2179 if "pandas" in str(type(args[0])): 2180 if "ytitle" not in kwargs: 2181 kwargs.update({"ytitle": args[0].name.replace("_", "_ ")}) 2182 x = np.linspace(0, len(args[0]), num=len(args[0])) 2183 y = np.asarray(args[0]).ravel() 2184 2185 elif utils.is_sequence(args[1]): 2186 # print('------------- case 3', 'plot(allx,ally)',str(type(args[0]))) 2187 if "pandas" in str(type(args[0])): 2188 if "xtitle" not in kwargs: 2189 kwargs.update({"xtitle": args[0].name.replace("_", "_ ")}) 2190 if "pandas" in str(type(args[1])): 2191 if "ytitle" not in kwargs: 2192 kwargs.update({"ytitle": args[1].name.replace("_", "_ ")}) 2193 x = np.asarray(args[0]).ravel() 2194 y = np.asarray(args[1]).ravel() 2195 2196 elif utils.is_sequence(args[0]) and utils.is_sequence(args[0][0]): 2197 # print('------------- case 4', 'plot([allx,ally])') 2198 x = np.asarray(args[0][0]).ravel() 2199 y = np.asarray(args[0][1]).ravel() 2200 2201 elif optidx == 2: 2202 # print('------------- case 5', 'plot(x,y)') 2203 x = np.asarray(args[0]).ravel() 2204 y = np.asarray(args[1]).ravel() 2205 2206 else: 2207 vedo.logger.error(f"plot(): Could not understand input arguments {args}") 2208 return None 2209 2210 if "polar" in mode: 2211 return _plot_polar(np.c_[x, y], **kwargs) 2212 2213 return PlotXY(np.c_[x, y], **kwargs) 2214 2215 2216def histogram(*args, **kwargs): 2217 """ 2218 Histogramming for 1D and 2D data arrays. 2219 2220 This is meant as a convenience function that creates the appropriate object 2221 based on the shape of the provided input data. 2222 2223 Use keyword `like=...` if you want to use the same format of a previously 2224 created Figure (useful when superimposing Figures) to make sure 2225 they are compatible and comparable. If they are not compatible 2226 you will receive an error message. 2227 2228 ------------------------------------------------------------------------- 2229 .. note:: default mode, for 1D arrays 2230 2231 Creates a `Histogram1D(Figure)` object. 2232 2233 Arguments: 2234 weights : (list) 2235 An array of weights, of the same shape as `data`. Each value in `data` 2236 only contributes its associated weight towards the bin count (instead of 1). 2237 bins : (int) 2238 number of bins 2239 vrange : (list) 2240 restrict the range of the histogram 2241 density : (bool) 2242 normalize the area to 1 by dividing by the nr of entries and bin size 2243 logscale : (bool) 2244 use logscale on y-axis 2245 fill : (bool) 2246 fill bars with solid color `c` 2247 gap : (float) 2248 leave a small space btw bars 2249 radius : (float) 2250 border radius of the top of the histogram bar. Default value is 0.1. 2251 texture : (str) 2252 url or path to an image to be used as texture for the bin 2253 outline : (bool) 2254 show outline of the bins 2255 errors : (bool) 2256 show error bars 2257 xtitle : (str) 2258 title for the x-axis, can also be set using `axes=dict(xtitle="my x axis")` 2259 ytitle : (str) 2260 title for the y-axis, can also be set using `axes=dict(ytitle="my y axis")` 2261 padding : (float, list) 2262 keep a padding space from the axes (as a fraction of the axis size). 2263 This can be a list of four numbers. 2264 aspect : (float) 2265 the desired aspect ratio of the histogram. Default is 4/3. 2266 grid : (bool) 2267 show the background grid for the axes, can also be set using `axes=dict(xygrid=True)` 2268 ztolerance : (float) 2269 a tolerance factor to superimpose objects (along the z-axis). 2270 2271 Examples: 2272 - [histo_1d_a.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_1d_a.py) 2273 - [histo_1d_b.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_1d_b.py) 2274 - [histo_1d_c.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_1d_c.py) 2275 - [histo_1d_d.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_1d_d.py) 2276 2277  2278 2279 2280 ------------------------------------------------------------------------- 2281 .. note:: default mode, for 2D arrays 2282 2283 Input data formats `[(x1,x2,..), (y1,y2,..)] or [(x1,y1), (x2,y2),..]` 2284 are both valid. 2285 2286 Arguments: 2287 bins : (list) 2288 binning as (nx, ny) 2289 weights : (list) 2290 array of weights to assign to each entry 2291 cmap : (str, lookuptable) 2292 color map name or look up table 2293 alpha : (float) 2294 opacity of the histogram 2295 gap : (float) 2296 separation between adjacent bins as a fraction for their size. 2297 Set gap=-1 to generate a quad surface. 2298 scalarbar : (bool) 2299 add a scalarbar to right of the histogram 2300 like : (Figure) 2301 grab and use the same format of the given Figure (for superimposing) 2302 xlim : (list) 2303 [x0, x1] range of interest. If left to None will automatically 2304 choose the minimum or the maximum of the data range. 2305 Data outside the range are completely ignored. 2306 ylim : (list) 2307 [y0, y1] range of interest. If left to None will automatically 2308 choose the minimum or the maximum of the data range. 2309 Data outside the range are completely ignored. 2310 aspect : (float) 2311 the desired aspect ratio of the figure. 2312 title : (str) 2313 title of the plot to appear on top. 2314 If left blank some statistics will be shown. 2315 xtitle : (str) 2316 x axis title 2317 ytitle : (str) 2318 y axis title 2319 ztitle : (str) 2320 title for the scalar bar 2321 ac : (str) 2322 axes color, additional keyword for Axes can also be added 2323 using e.g. `axes=dict(xygrid=True)` 2324 2325 Examples: 2326 - [histo_2d_a.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_2d_a.py) 2327 - [histo_2d_b.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_2d_b.py) 2328 2329  2330 2331 2332 ------------------------------------------------------------------------- 2333 .. note:: mode="3d" 2334 2335 If `mode='3d'`, build a 2D histogram as 3D bars from a list of x and y values. 2336 2337 Arguments: 2338 xtitle : (str) 2339 x axis title 2340 bins : (int) 2341 nr of bins for the smaller range in x or y 2342 vrange : (list) 2343 range in x and y in format `[(xmin,xmax), (ymin,ymax)]` 2344 norm : (float) 2345 sets a scaling factor for the z axis (frequency axis) 2346 fill : (bool) 2347 draw solid hexagons 2348 cmap : (str) 2349 color map name for elevation 2350 gap : (float) 2351 keep a internal empty gap between bins [0,1] 2352 zscale : (float) 2353 rescale the (already normalized) zaxis for visual convenience 2354 2355 Examples: 2356 - [histo_2d_b.py](https://github.com/marcomusy/vedo/tree/master/examples/examples/pyplot/histo_2d_b.py) 2357 2358 2359 ------------------------------------------------------------------------- 2360 .. note:: mode="hexbin" 2361 2362 If `mode='hexbin'`, build a hexagonal histogram from a list of x and y values. 2363 2364 Arguments: 2365 xtitle : (str) 2366 x axis title 2367 bins : (int) 2368 nr of bins for the smaller range in x or y 2369 vrange : (list) 2370 range in x and y in format `[(xmin,xmax), (ymin,ymax)]` 2371 norm : (float) 2372 sets a scaling factor for the z axis (frequency axis) 2373 fill : (bool) 2374 draw solid hexagons 2375 cmap : (str) 2376 color map name for elevation 2377 2378 Examples: 2379 - [histo_hexagonal.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_hexagonal.py) 2380 2381  2382 2383 2384 ------------------------------------------------------------------------- 2385 .. note:: mode="polar" 2386 2387 If `mode='polar'` assume input is polar coordinate system (rho, theta): 2388 2389 Arguments: 2390 weights : (list) 2391 Array of weights, of the same shape as the input. 2392 Each value only contributes its associated weight towards the bin count (instead of 1). 2393 title : (str) 2394 histogram title 2395 tsize : (float) 2396 title size 2397 bins : (int) 2398 number of bins in phi 2399 r1 : (float) 2400 inner radius 2401 r2 : (float) 2402 outer radius 2403 phigap : (float) 2404 gap angle btw 2 radial bars, in degrees 2405 rgap : (float) 2406 gap factor along radius of numeric angle labels 2407 lpos : (float) 2408 label gap factor along radius 2409 lsize : (float) 2410 label size 2411 c : (color) 2412 color of the histogram bars, can be a list of length `bins` 2413 bc : (color) 2414 color of the frame and labels 2415 alpha : (float) 2416 opacity of the frame 2417 cmap : (str) 2418 color map name 2419 deg : (bool) 2420 input array is in degrees 2421 vmin : (float) 2422 minimum value of the radial axis 2423 vmax : (float) 2424 maximum value of the radial axis 2425 labels : (list) 2426 list of labels, must be of length `bins` 2427 show_disc : (bool) 2428 show the outer ring axis 2429 nrays : (int) 2430 draw this number of axis rays (continuous and dashed) 2431 show_lines : (bool) 2432 show lines to the origin 2433 show_angles : (bool) 2434 show angular values 2435 show_errors : (bool) 2436 show error bars 2437 2438 Examples: 2439 - [histo_polar.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_polar.py) 2440 2441  2442 2443 2444 ------------------------------------------------------------------------- 2445 .. note:: mode="spheric" 2446 2447 If `mode='spheric'`, build a histogram from list of theta and phi values. 2448 2449 Arguments: 2450 rmax : (float) 2451 maximum radial elevation of bin 2452 res : (int) 2453 sphere resolution 2454 cmap : (str) 2455 color map name 2456 lw : (int) 2457 line width of the bin edges 2458 2459 Examples: 2460 - [histo_spheric.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_spheric.py) 2461 2462  2463 """ 2464 mode = kwargs.pop("mode", "") 2465 if len(args) == 2: # x, y 2466 2467 if "spher" in mode: 2468 return _histogram_spheric(args[0], args[1], **kwargs) 2469 2470 if "hex" in mode: 2471 return _histogram_hex_bin(args[0], args[1], **kwargs) 2472 2473 if "3d" in mode.lower(): 2474 return _histogram_quad_bin(args[0], args[1], **kwargs) 2475 2476 return Histogram2D(args[0], args[1], **kwargs) 2477 2478 elif len(args) == 1: 2479 2480 if isinstance(args[0], vedo.Volume): 2481 data = args[0].pointdata[0] 2482 elif isinstance(args[0], vedo.Points): 2483 pd0 = args[0].pointdata[0] 2484 if pd0: 2485 data = pd0.ravel() 2486 else: 2487 data = args[0].celldata[0].ravel() 2488 else: 2489 try: 2490 if "pandas" in str(type(args[0])): 2491 if "xtitle" not in kwargs: 2492 kwargs.update({"xtitle": args[0].name.replace("_", "_ ")}) 2493 except: 2494 pass 2495 data = np.asarray(args[0]) 2496 2497 if "spher" in mode: 2498 return _histogram_spheric(args[0][:, 0], args[0][:, 1], **kwargs) 2499 2500 if data.ndim == 1: 2501 if "polar" in mode: 2502 return _histogram_polar(data, **kwargs) 2503 return Histogram1D(data, **kwargs) 2504 2505 if "hex" in mode: 2506 return _histogram_hex_bin(args[0][:, 0], args[0][:, 1], **kwargs) 2507 2508 if "3d" in mode.lower(): 2509 return _histogram_quad_bin(args[0][:, 0], args[0][:, 1], **kwargs) 2510 2511 return Histogram2D(args[0], **kwargs) 2512 2513 vedo.logger.error(f"in histogram(): could not understand input {args[0]}") 2514 return None 2515 2516 2517def fit( 2518 points, deg=1, niter=0, nstd=3, xerrors=None, yerrors=None, vrange=None, res=250, lw=3, c="red4" 2519): 2520 """ 2521 Polynomial fitting with parameter error and error bands calculation. 2522 Errors bars in both x and y are supported. 2523 2524 Returns a `vedo.shapes.Line` object. 2525 2526 Additional information about the fitting output can be accessed with: 2527 2528 `fitd = fit(pts)` 2529 2530 - `fitd.coefficients` will contain the coefficients of the polynomial fit 2531 - `fitd.coefficient_errors`, errors on the fitting coefficients 2532 - `fitd.monte_carlo_coefficients`, fitting coefficient set from MC generation 2533 - `fitd.covariance_matrix`, covariance matrix as a numpy array 2534 - `fitd.reduced_chi2`, reduced chi-square of the fitting 2535 - `fitd.ndof`, number of degrees of freedom 2536 - `fitd.data_sigma`, mean data dispersion from the central fit assuming `Chi2=1` 2537 - `fitd.error_lines`, a `vedo.shapes.Line` object for the upper and lower error band 2538 - `fitd.error_band`, the `vedo.mesh.Mesh` object representing the error band 2539 2540 Errors on x and y can be specified. If left to `None` an estimate is made from 2541 the statistical spread of the dataset itself. Errors are always assumed gaussian. 2542 2543 Arguments: 2544 deg : (int) 2545 degree of the polynomial to be fitted 2546 niter : (int) 2547 number of monte-carlo iterations to compute error bands. 2548 If set to 0, return the simple least-squares fit with naive error estimation 2549 on coefficients only. A reasonable non-zero value to set is about 500, in 2550 this case *error_lines*, *error_band* and the other class attributes are filled 2551 nstd : (float) 2552 nr. of standard deviation to use for error calculation 2553 xerrors : (list) 2554 array of the same length of points with the errors on x 2555 yerrors : (list) 2556 array of the same length of points with the errors on y 2557 vrange : (list) 2558 specify the domain range of the fitting line 2559 (only affects visualization, but can be used to extrapolate the fit 2560 outside the data range) 2561 res : (int) 2562 resolution of the output fitted line and error lines 2563 2564 Examples: 2565 - [fit_polynomial1.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/fit_polynomial1.py) 2566 2567 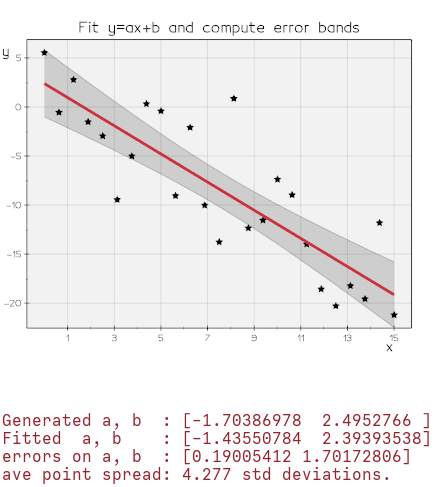 2568 """ 2569 if isinstance(points, vedo.pointcloud.Points): 2570 points = points.points() 2571 points = np.asarray(points) 2572 if len(points) == 2: # assume user is passing [x,y] 2573 points = np.c_[points[0], points[1]] 2574 x = points[:, 0] 2575 y = points[:, 1] # ignore z 2576 2577 n = len(x) 2578 ndof = n - deg - 1 2579 if vrange is not None: 2580 x0, x1 = vrange 2581 else: 2582 x0, x1 = np.min(x), np.max(x) 2583 if xerrors is not None: 2584 x0 -= xerrors[0] / 2 2585 x1 += xerrors[-1] / 2 2586 2587 tol = (x1 - x0) / 10000 2588 xr = np.linspace(x0, x1, res) 2589 2590 # project x errs on y 2591 if xerrors is not None: 2592 xerrors = np.asarray(xerrors) 2593 if yerrors is not None: 2594 yerrors = np.asarray(yerrors) 2595 w = 1.0 / yerrors 2596 coeffs = np.polyfit(x, y, deg, w=w, rcond=None) 2597 else: 2598 coeffs = np.polyfit(x, y, deg, rcond=None) 2599 # update yerrors, 1 bootstrap iteration is enough 2600 p1d = np.poly1d(coeffs) 2601 der = (p1d(x + tol) - p1d(x)) / tol 2602 yerrors = np.sqrt(yerrors * yerrors + np.power(der * xerrors, 2)) 2603 2604 if yerrors is not None: 2605 yerrors = np.asarray(yerrors) 2606 w = 1.0 / yerrors 2607 coeffs, V = np.polyfit(x, y, deg, w=w, rcond=None, cov=True) 2608 else: 2609 w = 1 2610 coeffs, V = np.polyfit(x, y, deg, rcond=None, cov=True) 2611 2612 p1d = np.poly1d(coeffs) 2613 theor = p1d(xr) 2614 fitl = shapes.Line(xr, theor, lw=lw, c=c).z(tol * 2) 2615 fitl.coefficients = coeffs 2616 fitl.covariance_matrix = V 2617 residuals2_sum = np.sum(np.power(p1d(x) - y, 2)) / ndof 2618 sigma = np.sqrt(residuals2_sum) 2619 fitl.reduced_chi2 = np.sum(np.power((p1d(x) - y) * w, 2)) / ndof 2620 fitl.ndof = ndof 2621 fitl.data_sigma = sigma # worked out from data using chi2=1 hypo 2622 fitl.name = "LinearPolynomialFit" 2623 2624 if not niter: 2625 fitl.coefficient_errors = np.sqrt(np.diag(V)) 2626 return fitl ################################ 2627 2628 if yerrors is not None: 2629 sigma = yerrors 2630 else: 2631 w = None 2632 fitl.reduced_chi2 = 1 2633 2634 Theors, all_coeffs = [], [] 2635 for i in range(niter): 2636 noise = np.random.randn(n) * sigma 2637 coeffs = np.polyfit(x, y + noise, deg, w=w, rcond=None) 2638 all_coeffs.append(coeffs) 2639 P1d = np.poly1d(coeffs) 2640 Theor = P1d(xr) 2641 Theors.append(Theor) 2642 all_coeffs = np.array(all_coeffs) 2643 fitl.monte_carlo_coefficients = all_coeffs 2644 2645 stds = np.std(Theors, axis=0) 2646 fitl.coefficient_errors = np.std(all_coeffs, axis=0) 2647 2648 # check distributions on the fly 2649 # for i in range(deg+1): 2650 # histogram(all_coeffs[:,i],title='par'+str(i)).show(new=1) 2651 # histogram(all_coeffs[:,0], all_coeffs[:,1], 2652 # xtitle='param0', ytitle='param1',scalarbar=1).show(new=1) 2653 # histogram(all_coeffs[:,1], all_coeffs[:,2], 2654 # xtitle='param1', ytitle='param2').show(new=1) 2655 # histogram(all_coeffs[:,0], all_coeffs[:,2], 2656 # xtitle='param0', ytitle='param2').show(new=1) 2657 2658 error_lines = [] 2659 for i in [nstd, -nstd]: 2660 el = shapes.Line(xr, theor + stds * i, lw=1, alpha=0.2, c="k").z(tol) 2661 error_lines.append(el) 2662 el.name = "ErrorLine for sigma=" + str(i) 2663 2664 fitl.error_lines = error_lines 2665 l1 = error_lines[0].points().tolist() 2666 cband = l1 + list(reversed(error_lines[1].points().tolist())) + [l1[0]] 2667 fitl.error_band = shapes.Line(cband).triangulate().lw(0).c("k", 0.15) 2668 fitl.error_band.name = "PolynomialFitErrorBand" 2669 return fitl 2670 2671 2672def _plot_fxy( 2673 z, 2674 xlim=(0, 3), 2675 ylim=(0, 3), 2676 zlim=(None, None), 2677 show_nan=True, 2678 zlevels=10, 2679 c=None, 2680 bc="aqua", 2681 alpha=1, 2682 texture="", 2683 bins=(100, 100), 2684 axes=True, 2685): 2686 if c is not None: 2687 texture = None # disable 2688 2689 ps = vtk.vtkPlaneSource() 2690 ps.SetResolution(bins[0], bins[1]) 2691 ps.SetNormal([0, 0, 1]) 2692 ps.Update() 2693 poly = ps.GetOutput() 2694 dx = xlim[1] - xlim[0] 2695 dy = ylim[1] - ylim[0] 2696 todel, nans = [], [] 2697 2698 for i in range(poly.GetNumberOfPoints()): 2699 px, py, _ = poly.GetPoint(i) 2700 xv = (px + 0.5) * dx + xlim[0] 2701 yv = (py + 0.5) * dy + ylim[0] 2702 try: 2703 with warnings.catch_warnings(): 2704 warnings.simplefilter("ignore") 2705 zv = z(xv, yv) 2706 if np.isnan(zv) or np.isinf(zv) or np.iscomplex(zv): 2707 zv = 0 2708 todel.append(i) 2709 nans.append([xv, yv, 0]) 2710 except: 2711 zv = 0 2712 todel.append(i) 2713 nans.append([xv, yv, 0]) 2714 poly.GetPoints().SetPoint(i, [xv, yv, zv]) 2715 2716 if todel: 2717 cellIds = vtk.vtkIdList() 2718 poly.BuildLinks() 2719 for i in todel: 2720 poly.GetPointCells(i, cellIds) 2721 for j in range(cellIds.GetNumberOfIds()): 2722 poly.DeleteCell(cellIds.GetId(j)) # flag cell 2723 poly.RemoveDeletedCells() 2724 cl = vtk.vtkCleanPolyData() 2725 cl.SetInputData(poly) 2726 cl.Update() 2727 poly = cl.GetOutput() 2728 2729 if not poly.GetNumberOfPoints(): 2730 vedo.logger.error("function is not real in the domain") 2731 return None 2732 2733 if zlim[0]: 2734 tmpact1 = Mesh(poly) 2735 a = tmpact1.cut_with_plane((0, 0, zlim[0]), (0, 0, 1)) 2736 poly = a.polydata() 2737 if zlim[1]: 2738 tmpact2 = Mesh(poly) 2739 a = tmpact2.cut_with_plane((0, 0, zlim[1]), (0, 0, -1)) 2740 poly = a.polydata() 2741 2742 cmap = "" 2743 if c in colors.cmaps_names: 2744 cmap = c 2745 c = None 2746 bc = None 2747 2748 mesh = Mesh(poly, c, alpha).compute_normals().lighting("plastic") 2749 2750 if cmap: 2751 mesh.compute_elevation().cmap(cmap) 2752 if bc: 2753 mesh.bc(bc) 2754 if texture: 2755 mesh.texture(texture) 2756 2757 acts = [mesh] 2758 if zlevels: 2759 elevation = vtk.vtkElevationFilter() 2760 elevation.SetInputData(poly) 2761 bounds = poly.GetBounds() 2762 elevation.SetLowPoint(0, 0, bounds[4]) 2763 elevation.SetHighPoint(0, 0, bounds[5]) 2764 elevation.Update() 2765 bcf = vtk.vtkBandedPolyDataContourFilter() 2766 bcf.SetInputData(elevation.GetOutput()) 2767 bcf.SetScalarModeToValue() 2768 bcf.GenerateContourEdgesOn() 2769 bcf.GenerateValues(zlevels, elevation.GetScalarRange()) 2770 bcf.Update() 2771 zpoly = bcf.GetContourEdgesOutput() 2772 zbandsact = Mesh(zpoly, "k", alpha).lw(1).lighting("off") 2773 zbandsact.mapper().SetResolveCoincidentTopologyToPolygonOffset() 2774 acts.append(zbandsact) 2775 2776 if show_nan and todel: 2777 bb = mesh.GetBounds() 2778 if bb[4] <= 0 and bb[5] >= 0: 2779 zm = 0.0 2780 else: 2781 zm = (bb[4] + bb[5]) / 2 2782 nans = np.array(nans) + [0, 0, zm] 2783 nansact = shapes.Points(nans, r=2, c="red5", alpha=alpha) 2784 nansact.GetProperty().RenderPointsAsSpheresOff() 2785 acts.append(nansact) 2786 2787 if isinstance(axes, dict): 2788 axs = addons.Axes(mesh, **axes) 2789 acts.append(axs) 2790 elif axes: 2791 axs = addons.Axes(mesh) 2792 acts.append(axs) 2793 2794 assem = Assembly(acts) 2795 assem.name = "PlotFxy" 2796 return assem 2797 2798 2799def _plot_fz( 2800 z, 2801 x=(-1, 1), 2802 y=(-1, 1), 2803 zlimits=(None, None), 2804 cmap="PiYG", 2805 alpha=1, 2806 lw=0.1, 2807 bins=(75, 75), 2808 axes=True, 2809): 2810 ps = vtk.vtkPlaneSource() 2811 ps.SetResolution(bins[0], bins[1]) 2812 ps.SetNormal([0, 0, 1]) 2813 ps.Update() 2814 poly = ps.GetOutput() 2815 dx = x[1] - x[0] 2816 dy = y[1] - y[0] 2817 2818 arrImg = [] 2819 for i in range(poly.GetNumberOfPoints()): 2820 px, py, _ = poly.GetPoint(i) 2821 xv = (px + 0.5) * dx + x[0] 2822 yv = (py + 0.5) * dy + y[0] 2823 try: 2824 zv = z(complex(xv), complex(yv)) 2825 except: 2826 zv = 0 2827 poly.GetPoints().SetPoint(i, [xv, yv, np.real(zv)]) 2828 arrImg.append(np.imag(zv)) 2829 2830 mesh = Mesh(poly, alpha).lighting("plastic") 2831 v = max(abs(np.min(arrImg)), abs(np.max(arrImg))) 2832 mesh.cmap(cmap, arrImg, vmin=-v, vmax=v) 2833 mesh.compute_normals().lw(lw) 2834 2835 if zlimits[0]: 2836 mesh.cut_with_plane((0, 0, zlimits[0]), (0, 0, 1)) 2837 if zlimits[1]: 2838 mesh.cut_with_plane((0, 0, zlimits[1]), (0, 0, -1)) 2839 2840 acts = [mesh] 2841 if axes: 2842 axs = addons.Axes(mesh, ztitle="Real part") 2843 acts.append(axs) 2844 asse = Assembly(acts) 2845 asse.name = "PlotFz" 2846 if isinstance(z, str): 2847 asse.name += " " + z 2848 return asse 2849 2850 2851def _plot_polar( 2852 rphi, 2853 title="", 2854 tsize=0.1, 2855 lsize=0.05, 2856 r1=0, 2857 r2=1, 2858 c="blue", 2859 bc="k", 2860 alpha=1, 2861 ps=5, 2862 lw=3, 2863 deg=False, 2864 vmax=None, 2865 fill=False, 2866 splined=False, 2867 nrays=8, 2868 show_disc=True, 2869 show_lines=True, 2870 show_angles=True, 2871): 2872 if len(rphi) == 2: 2873 rphi = np.stack((rphi[0], rphi[1]), axis=1) 2874 2875 rphi = np.array(rphi, dtype=float) 2876 thetas = rphi[:, 0] 2877 radii = rphi[:, 1] 2878 2879 k = 180 / np.pi 2880 if deg: 2881 thetas = np.array(thetas, dtype=float) / k 2882 2883 vals = [] 2884 for v in thetas: # normalize range 2885 t = np.arctan2(np.sin(v), np.cos(v)) 2886 if t < 0: 2887 t += 2 * np.pi 2888 vals.append(t) 2889 thetas = np.array(vals, dtype=float) 2890 2891 if vmax is None: 2892 vmax = np.max(radii) 2893 2894 angles = [] 2895 points = [] 2896 for t, r in zip(thetas, radii): 2897 r = r / vmax * r2 + r1 2898 ct, st = np.cos(t), np.sin(t) 2899 points.append([r * ct, r * st, 0]) 2900 p0 = points[0] 2901 points.append(p0) 2902 2903 r2e = r1 + r2 2904 lines = None 2905 if splined: 2906 lines = shapes.KSpline(points, closed=True) 2907 lines.c(c).lw(lw).alpha(alpha) 2908 elif lw: 2909 lines = shapes.Line(points) 2910 lines.c(c).lw(lw).alpha(alpha) 2911 2912 points.pop() 2913 2914 ptsact = None 2915 if ps: 2916 ptsact = shapes.Points(points, r=ps, c=c, alpha=alpha) 2917 2918 filling = None 2919 if fill and lw: 2920 faces = [] 2921 coords = [[0, 0, 0]] + lines.points().tolist() 2922 for i in range(1, lines.npoints): 2923 faces.append([0, i, i + 1]) 2924 filling = Mesh([coords, faces]).c(c).alpha(alpha) 2925 2926 back = None 2927 back2 = None 2928 if show_disc: 2929 back = shapes.Disc(r1=r2e, r2=r2e * 1.01, c=bc, res=(1, 360)) 2930 back.z(-0.01).lighting("off").alpha(alpha) 2931 back2 = shapes.Disc(r1=r2e / 2, r2=r2e / 2 * 1.005, c=bc, res=(1, 360)) 2932 back2.z(-0.01).lighting("off").alpha(alpha) 2933 2934 ti = None 2935 if title: 2936 ti = shapes.Text3D(title, (0, 0, 0), s=tsize, depth=0, justify="top-center") 2937 ti.pos(0, -r2e * 1.15, 0.01) 2938 2939 rays = [] 2940 if show_disc: 2941 rgap = 0.05 2942 for t in np.linspace(0, 2 * np.pi, num=nrays, endpoint=False): 2943 ct, st = np.cos(t), np.sin(t) 2944 if show_lines: 2945 l = shapes.Line((0, 0, -0.01), (r2e * ct * 1.03, r2e * st * 1.03, -0.01)) 2946 rays.append(l) 2947 ct2, st2 = np.cos(t + np.pi / nrays), np.sin(t + np.pi / nrays) 2948 lm = shapes.DashedLine((0, 0, -0.01), (r2e * ct2, r2e * st2, -0.01), spacing=0.25) 2949 rays.append(lm) 2950 elif show_angles: # just the ticks 2951 l = shapes.Line( 2952 (r2e * ct * 0.98, r2e * st * 0.98, -0.01), 2953 (r2e * ct * 1.03, r2e * st * 1.03, -0.01), 2954 ) 2955 if show_angles: 2956 if 0 <= t < np.pi / 2: 2957 ju = "bottom-left" 2958 elif t == np.pi / 2: 2959 ju = "bottom-center" 2960 elif np.pi / 2 < t <= np.pi: 2961 ju = "bottom-right" 2962 elif np.pi < t < np.pi * 3 / 2: 2963 ju = "top-right" 2964 elif t == np.pi * 3 / 2: 2965 ju = "top-center" 2966 else: 2967 ju = "top-left" 2968 a = shapes.Text3D(int(t * k), pos=(0, 0, 0), s=lsize, depth=0, justify=ju) 2969 a.pos(r2e * ct * (1 + rgap), r2e * st * (1 + rgap), -0.01) 2970 angles.append(a) 2971 2972 mrg = merge(back, back2, angles, rays, ti) 2973 if mrg: 2974 mrg.color(bc).alpha(alpha).lighting("off") 2975 rh = Assembly([lines, ptsact, filling] + [mrg]) 2976 rh.base = np.array([0, 0, 0], dtype=float) 2977 rh.top = np.array([0, 0, 1], dtype=float) 2978 rh.name = "PlotPolar" 2979 return rh 2980 2981 2982def _plot_spheric(rfunc, normalize=True, res=33, scalarbar=True, c="grey", alpha=0.05, cmap="jet"): 2983 sg = shapes.Sphere(res=res, quads=True) 2984 sg.alpha(alpha).c(c).wireframe() 2985 2986 cgpts = sg.points() 2987 r, theta, phi = utils.cart2spher(*cgpts.T) 2988 2989 newr, inans = [], [] 2990 for i in range(len(r)): 2991 try: 2992 ri = rfunc(theta[i], phi[i]) 2993 if np.isnan(ri): 2994 inans.append(i) 2995 newr.append(1) 2996 else: 2997 newr.append(ri) 2998 except: 2999 inans.append(i) 3000 newr.append(1) 3001 3002 newr = np.array(newr, dtype=float) 3003 if normalize: 3004 newr = newr / np.max(newr) 3005 newr[inans] = 1 3006 3007 nanpts = [] 3008 if inans: 3009 redpts = utils.spher2cart(newr[inans], theta[inans], phi[inans]) 3010 nanpts.append(shapes.Points(redpts, r=4, c="r")) 3011 3012 pts = utils.spher2cart(newr, theta, phi) 3013 3014 ssurf = sg.clone().points(pts) 3015 if inans: 3016 ssurf.delete_cells_by_point_index(inans) 3017 3018 ssurf.alpha(1).wireframe(0).lw(0.1) 3019 3020 ssurf.cmap(cmap, newr) 3021 ssurf.compute_normals() 3022 3023 if scalarbar: 3024 xm = np.max([np.max(pts[0]), 1]) 3025 ym = np.max([np.abs(np.max(pts[1])), 1]) 3026 ssurf.mapper().SetScalarRange(np.min(newr), np.max(newr)) 3027 sb3d = ssurf.add_scalarbar3d(size=(xm * 0.07, ym), c="k").scalarbar 3028 sb3d.rotate_x(90).pos(xm * 1.1, 0, -0.5) 3029 else: 3030 sb3d = None 3031 3032 sg.pickable(False) 3033 asse = Assembly([ssurf, sg] + nanpts + [sb3d]) 3034 asse.name = "PlotSpheric" 3035 return asse 3036 3037 3038def _histogram_quad_bin(x, y, **kwargs): 3039 # generate a histogram with 3D bars 3040 # 3041 histo = Histogram2D(x, y, **kwargs) 3042 3043 gap = kwargs.pop("gap", 0) 3044 zscale = kwargs.pop("zscale", 1) 3045 cmap = kwargs.pop("cmap", "Blues_r") 3046 3047 gr = histo.actors[2] 3048 d = gr.diagonal_size() 3049 tol = d / 1_000_000 # tolerance 3050 if gap >= 0: 3051 gr.shrink(1 - gap - tol) 3052 gr.map_cells_to_points() 3053 3054 faces = np.array(gr.faces()) 3055 s = 1 / histo.entries * len(faces) * zscale 3056 zvals = gr.pointdata["Scalars"] * s 3057 3058 pts1 = gr.points() 3059 pts2 = np.copy(pts1) 3060 pts2[:, 2] = zvals + tol 3061 newpts = np.vstack([pts1, pts2]) 3062 newzvals = np.hstack([zvals, zvals]) / s 3063 3064 n = pts1.shape[0] 3065 newfaces = [] 3066 for f in faces: 3067 f0, f1, f2, f3 = f 3068 f0n, f1n, f2n, f3n = f + n 3069 newfaces.extend( 3070 [ 3071 [f0, f1, f2, f3], 3072 [f0n, f1n, f2n, f3n], 3073 [f0, f1, f1n, f0n], 3074 [f1, f2, f2n, f1n], 3075 [f2, f3, f3n, f2n], 3076 [f3, f0, f0n, f3n], 3077 ] 3078 ) 3079 3080 msh = Mesh([newpts, newfaces]).pickable(False) 3081 msh.cmap(cmap, newzvals, name="Frequency") 3082 msh.lw(1).lighting("ambient") 3083 3084 histo.actors[2] = msh 3085 histo.RemovePart(gr) 3086 histo.AddPart(msh) 3087 return histo 3088 3089 3090def _histogram_hex_bin( 3091 xvalues, yvalues, bins=12, norm=1, fill=True, c=None, cmap="terrain_r", alpha=1 3092): 3093 xmin, xmax = np.min(xvalues), np.max(xvalues) 3094 ymin, ymax = np.min(yvalues), np.max(yvalues) 3095 dx, dy = xmax - xmin, ymax - ymin 3096 3097 if utils.is_sequence(bins): 3098 n, m = bins 3099 else: 3100 if xmax - xmin < ymax - ymin: 3101 n = bins 3102 m = np.rint(dy / dx * n / 1.2 + 0.5).astype(int) 3103 else: 3104 m = bins 3105 n = np.rint(dx / dy * m * 1.2 + 0.5).astype(int) 3106 3107 src = vtk.vtkPointSource() 3108 src.SetNumberOfPoints(len(xvalues)) 3109 src.Update() 3110 poly = src.GetOutput() 3111 3112 values = np.stack((xvalues, yvalues), axis=1) 3113 zs = [[0.0]] * len(values) 3114 values = np.append(values, zs, axis=1) 3115 3116 poly.GetPoints().SetData(utils.numpy2vtk(values, dtype=np.float32)) 3117 cloud = Mesh(poly) 3118 3119 col = None 3120 if c is not None: 3121 col = colors.get_color(c) 3122 3123 hexs, binmax = [], 0 3124 ki, kj = 1.33, 1.12 3125 r = 0.47 / n * 1.2 * dx 3126 for i in range(n + 3): 3127 for j in range(m + 2): 3128 cyl = vtk.vtkCylinderSource() 3129 cyl.SetResolution(6) 3130 cyl.CappingOn() 3131 cyl.SetRadius(0.5) 3132 cyl.SetHeight(0.1) 3133 cyl.Update() 3134 t = vtk.vtkTransform() 3135 if not i % 2: 3136 p = (i / ki, j / kj, 0) 3137 else: 3138 p = (i / ki, j / kj + 0.45, 0) 3139 q = (p[0] / n * 1.2 * dx + xmin, p[1] / m * dy + ymin, 0) 3140 ids = cloud.closest_point(q, radius=r, return_cell_id=True) 3141 ne = len(ids) 3142 if fill: 3143 t.Translate(p[0], p[1], ne / 2) 3144 t.Scale(1, 1, ne * 10) 3145 else: 3146 t.Translate(p[0], p[1], ne) 3147 t.RotateX(90) # put it along Z 3148 tf = vtk.vtkTransformPolyDataFilter() 3149 tf.SetInputData(cyl.GetOutput()) 3150 tf.SetTransform(t) 3151 tf.Update() 3152 if c is None: 3153 col = i 3154 h = Mesh(tf.GetOutput(), c=col, alpha=alpha).flat() 3155 h.lighting("plastic") 3156 h.PickableOff() 3157 hexs.append(h) 3158 if ne > binmax: 3159 binmax = ne 3160 3161 if cmap is not None: 3162 for h in hexs: 3163 z = h.GetBounds()[5] 3164 col = colors.color_map(z, cmap, 0, binmax) 3165 h.color(col) 3166 3167 asse = Assembly(hexs) 3168 asse.SetScale(1.2 / n * dx, 1 / m * dy, norm / binmax * (dx + dy) / 4) 3169 asse.SetPosition(xmin, ymin, 0) 3170 asse.base = np.array([0, 0, 0], dtype=float) 3171 asse.top = np.array([0, 0, 1], dtype=float) 3172 asse.name = "HistogramHexBin" 3173 return asse 3174 3175 3176def _histogram_polar( 3177 values, 3178 weights=None, 3179 title="", 3180 tsize=0.1, 3181 bins=16, 3182 r1=0.25, 3183 r2=1, 3184 phigap=0.5, 3185 rgap=0.05, 3186 lpos=1, 3187 lsize=0.04, 3188 c="grey", 3189 bc="k", 3190 alpha=1, 3191 cmap=None, 3192 deg=False, 3193 vmin=None, 3194 vmax=None, 3195 labels=(), 3196 show_disc=True, 3197 nrays=8, 3198 show_lines=True, 3199 show_angles=True, 3200 show_errors=False, 3201): 3202 k = 180 / np.pi 3203 if deg: 3204 values = np.array(values, dtype=float) / k 3205 else: 3206 values = np.array(values, dtype=float) 3207 3208 vals = [] 3209 for v in values: # normalize range 3210 t = np.arctan2(np.sin(v), np.cos(v)) 3211 if t < 0: 3212 t += 2 * np.pi 3213 vals.append(t + 0.00001) 3214 3215 histodata, edges = np.histogram(vals, weights=weights, bins=bins, range=(0, 2 * np.pi)) 3216 3217 thetas = [] 3218 for i in range(bins): 3219 thetas.append((edges[i] + edges[i + 1]) / 2) 3220 3221 if vmin is None: 3222 vmin = np.min(histodata) 3223 if vmax is None: 3224 vmax = np.max(histodata) 3225 3226 errors = np.sqrt(histodata) 3227 r2e = r1 + r2 3228 if show_errors: 3229 r2e += np.max(errors) / vmax * 1.5 3230 3231 back = None 3232 if show_disc: 3233 back = shapes.Disc(r1=r2e, r2=r2e * 1.01, c=bc, res=(1, 360)) 3234 back.z(-0.01) 3235 3236 slices = [] 3237 lines = [] 3238 angles = [] 3239 errbars = [] 3240 3241 for i, t in enumerate(thetas): 3242 r = histodata[i] / vmax * r2 3243 d = shapes.Disc((0, 0, 0), r1, r1 + r, res=(1, 360)) 3244 delta = np.pi / bins - np.pi / 2 - phigap / k 3245 d.cut_with_plane(normal=(np.cos(t + delta), np.sin(t + delta), 0)) 3246 d.cut_with_plane(normal=(np.cos(t - delta), np.sin(t - delta), 0)) 3247 if cmap is not None: 3248 cslice = colors.color_map(histodata[i], cmap, vmin, vmax) 3249 d.color(cslice) 3250 else: 3251 if c is None: 3252 d.color(i) 3253 elif utils.is_sequence(c) and len(c) == bins: 3254 d.color(c[i]) 3255 else: 3256 d.color(c) 3257 d.alpha(alpha).lighting("off") 3258 slices.append(d) 3259 3260 ct, st = np.cos(t), np.sin(t) 3261 3262 if show_errors: 3263 show_lines = False 3264 err = np.sqrt(histodata[i]) / vmax * r2 3265 errl = shapes.Line( 3266 ((r1 + r - err) * ct, (r1 + r - err) * st, 0.01), 3267 ((r1 + r + err) * ct, (r1 + r + err) * st, 0.01), 3268 ) 3269 errl.alpha(alpha).lw(3).color(bc) 3270 errbars.append(errl) 3271 3272 labs = [] 3273 rays = [] 3274 if show_disc: 3275 outerdisc = shapes.Disc(r1=r2e, r2=r2e * 1.01, c=bc, res=(1, 360)) 3276 outerdisc.z(-0.01) 3277 innerdisc = shapes.Disc(r1=r2e / 2, r2=r2e / 2 * 1.005, c=bc, res=(1, 360)) 3278 innerdisc.z(-0.01) 3279 rays.append(outerdisc) 3280 rays.append(innerdisc) 3281 3282 rgap = 0.05 3283 for t in np.linspace(0, 2 * np.pi, num=nrays, endpoint=False): 3284 ct, st = np.cos(t), np.sin(t) 3285 if show_lines: 3286 l = shapes.Line((0, 0, -0.01), (r2e * ct * 1.03, r2e * st * 1.03, -0.01)) 3287 rays.append(l) 3288 ct2, st2 = np.cos(t + np.pi / nrays), np.sin(t + np.pi / nrays) 3289 lm = shapes.DashedLine((0, 0, -0.01), (r2e * ct2, r2e * st2, -0.01), spacing=0.25) 3290 rays.append(lm) 3291 elif show_angles: # just the ticks 3292 l = shapes.Line( 3293 (r2e * ct * 0.98, r2e * st * 0.98, -0.01), 3294 (r2e * ct * 1.03, r2e * st * 1.03, -0.01), 3295 ) 3296 if show_angles: 3297 if 0 <= t < np.pi / 2: 3298 ju = "bottom-left" 3299 elif t == np.pi / 2: 3300 ju = "bottom-center" 3301 elif np.pi / 2 < t <= np.pi: 3302 ju = "bottom-right" 3303 elif np.pi < t < np.pi * 3 / 2: 3304 ju = "top-right" 3305 elif t == np.pi * 3 / 2: 3306 ju = "top-center" 3307 else: 3308 ju = "top-left" 3309 a = shapes.Text3D(int(t * k), pos=(0, 0, 0), s=lsize, depth=0, justify=ju) 3310 a.pos(r2e * ct * (1 + rgap), r2e * st * (1 + rgap), -0.01) 3311 angles.append(a) 3312 3313 ti = None 3314 if title: 3315 ti = shapes.Text3D(title, (0, 0, 0), s=tsize, depth=0, justify="top-center") 3316 ti.pos(0, -r2e * 1.15, 0.01) 3317 3318 for i, t in enumerate(thetas): 3319 if i < len(labels): 3320 lab = shapes.Text3D( 3321 labels[i], (0, 0, 0), s=lsize, depth=0, justify="center" # font="VTK", 3322 ) 3323 lab.pos( 3324 r2e * np.cos(t) * (1 + rgap) * lpos / 2, 3325 r2e * np.sin(t) * (1 + rgap) * lpos / 2, 3326 0.01, 3327 ) 3328 labs.append(lab) 3329 3330 mrg = merge(lines, angles, rays, ti, labs) 3331 if mrg: 3332 mrg.color(bc).lighting("off") 3333 3334 acts = slices + errbars + [mrg] 3335 asse = Assembly(acts) 3336 asse.frequencies = histodata 3337 asse.bins = edges 3338 asse.name = "HistogramPolar" 3339 return asse 3340 3341 3342def _histogram_spheric(thetavalues, phivalues, rmax=1.2, res=8, cmap="rainbow", gap=0.1): 3343 3344 x, y, z = utils.spher2cart(np.ones_like(thetavalues) * 1.1, thetavalues, phivalues) 3345 ptsvals = np.c_[x, y, z] 3346 3347 sg = shapes.Sphere(res=res, quads=True).shrink(1 - gap) 3348 sgfaces = sg.faces() 3349 sgpts = sg.points() 3350 3351 cntrs = sg.cell_centers() 3352 counts = np.zeros(len(cntrs)) 3353 for p in ptsvals: 3354 cell = sg.closest_point(p, return_cell_id=True) 3355 counts[cell] += 1 3356 acounts = np.array(counts, dtype=float) 3357 counts *= (rmax - 1) / np.max(counts) 3358 3359 for cell, cn in enumerate(counts): 3360 if not cn: 3361 continue 3362 fs = sgfaces[cell] 3363 pts = sgpts[fs] 3364 _, t1, p1 = utils.cart2spher(pts[:, 0], pts[:, 1], pts[:, 2]) 3365 x, y, z = utils.spher2cart(1 + cn, t1, p1) 3366 sgpts[fs] = np.c_[x, y, z] 3367 3368 sg.points(sgpts) 3369 sg.cmap(cmap, acounts, on="cells") 3370 vals = sg.celldata["Scalars"] 3371 3372 faces = sg.faces() 3373 points = sg.points().tolist() + [[0.0, 0.0, 0.0]] 3374 lp = len(points) - 1 3375 newfaces = [] 3376 newvals = [] 3377 for i, f in enumerate(faces): 3378 p0, p1, p2, p3 = f 3379 newfaces.append(f) 3380 newfaces.append([p0, lp, p1]) 3381 newfaces.append([p1, lp, p2]) 3382 newfaces.append([p2, lp, p3]) 3383 newfaces.append([p3, lp, p0]) 3384 for _ in range(5): 3385 newvals.append(vals[i]) 3386 3387 newsg = Mesh([points, newfaces]).cmap(cmap, newvals, on="cells") 3388 newsg.compute_normals().flat() 3389 newsg.name = "HistogramSpheric" 3390 return newsg 3391 3392 3393def donut( 3394 fractions, 3395 title="", 3396 tsize=0.3, 3397 r1=1.7, 3398 r2=1, 3399 phigap=0, 3400 lpos=0.8, 3401 lsize=0.15, 3402 c=None, 3403 bc="k", 3404 alpha=1, 3405 labels=(), 3406 show_disc=False, 3407): 3408 """ 3409 Donut plot or pie chart. 3410 3411 Arguments: 3412 title : (str) 3413 plot title 3414 tsize : (float) 3415 title size 3416 r1 : (float) inner radius 3417 r2 : (float) 3418 outer radius, starting from r1 3419 phigap : (float) 3420 gap angle btw 2 radial bars, in degrees 3421 lpos : (float) 3422 label gap factor along radius 3423 lsize : (float) 3424 label size 3425 c : (color) 3426 color of the plot slices 3427 bc : (color) 3428 color of the disc frame 3429 alpha : (float) 3430 opacity of the disc frame 3431 labels : (list) 3432 list of labels 3433 show_disc : (bool) 3434 show the outer ring axis 3435 3436 Examples: 3437 - [donut.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/donut.py) 3438 3439  3440 """ 3441 fractions = np.array(fractions, dtype=float) 3442 angles = np.add.accumulate(2 * np.pi * fractions) 3443 angles[-1] = 2 * np.pi 3444 if angles[-2] > 2 * np.pi: 3445 print("Error in donut(): fractions must sum to 1.") 3446 raise RuntimeError 3447 3448 cols = [] 3449 for i, th in enumerate(np.linspace(0, 2 * np.pi, 360, endpoint=False)): 3450 for ia, a in enumerate(angles): 3451 if th < a: 3452 cols.append(c[ia]) 3453 break 3454 labs = () 3455 if labels: 3456 angles = np.concatenate([[0], angles]) 3457 labs = [""] * 360 3458 for i in range(len(labels)): 3459 a = (angles[i + 1] + angles[i]) / 2 3460 j = int(a / np.pi * 180) 3461 labs[j] = labels[i] 3462 3463 data = np.linspace(0, 2 * np.pi, 360, endpoint=False) + 0.005 3464 dn = _histogram_polar( 3465 data, 3466 title=title, 3467 bins=360, 3468 r1=r1, 3469 r2=r2, 3470 phigap=phigap, 3471 lpos=lpos, 3472 lsize=lsize, 3473 tsize=tsize, 3474 c=cols, 3475 bc=bc, 3476 alpha=alpha, 3477 vmin=0, 3478 vmax=1, 3479 labels=labs, 3480 show_disc=show_disc, 3481 show_lines=0, 3482 show_angles=0, 3483 show_errors=0, 3484 ) 3485 dn.name = "Donut" 3486 return dn 3487 3488 3489def violin( 3490 values, 3491 bins=10, 3492 vlim=None, 3493 x=0, 3494 width=3, 3495 splined=True, 3496 fill=True, 3497 c="violet", 3498 alpha=1, 3499 outline=True, 3500 centerline=True, 3501 lc="darkorchid", 3502 lw=3, 3503): 3504 """ 3505 Violin style histogram. 3506 3507 Arguments: 3508 bins : (int) 3509 number of bins 3510 vlim : (list) 3511 input value limits. Crop values outside range 3512 x : (float) 3513 x-position of the violin axis 3514 width : (float) 3515 width factor of the normalized distribution 3516 splined : (bool) 3517 spline the outline 3518 fill : (bool) 3519 fill violin with solid color 3520 outline : (bool) 3521 add the distribution outline 3522 centerline : (bool) 3523 add the vertical centerline at x 3524 lc : (color) 3525 line color 3526 3527 Examples: 3528 - [histo_violin.py](https://github.com/marcomusy/vedo/tree/master/examples/examples/pyplot/histo_violin.py) 3529 3530  3531 """ 3532 fs, edges = np.histogram(values, bins=bins, range=vlim) 3533 mine, maxe = np.min(edges), np.max(edges) 3534 fs = fs.astype(float) / len(values) * width 3535 3536 rs = [] 3537 3538 if splined: 3539 lnl, lnr = [(0, edges[0], 0)], [(0, edges[0], 0)] 3540 for i in range(bins): 3541 xc = (edges[i] + edges[i + 1]) / 2 3542 yc = fs[i] 3543 lnl.append([-yc, xc, 0]) 3544 lnr.append([yc, xc, 0]) 3545 lnl.append((0, edges[-1], 0)) 3546 lnr.append((0, edges[-1], 0)) 3547 spl = shapes.KSpline(lnl).x(x) 3548 spr = shapes.KSpline(lnr).x(x) 3549 spl.color(lc).alpha(alpha).lw(lw) 3550 spr.color(lc).alpha(alpha).lw(lw) 3551 if outline: 3552 rs.append(spl) 3553 rs.append(spr) 3554 if fill: 3555 rb = shapes.Ribbon(spl, spr, c=c, alpha=alpha).lighting("off") 3556 rs.append(rb) 3557 3558 else: 3559 lns1 = [[0, mine, 0]] 3560 for i in range(bins): 3561 lns1.append([fs[i], edges[i], 0]) 3562 lns1.append([fs[i], edges[i + 1], 0]) 3563 lns1.append([0, maxe, 0]) 3564 3565 lns2 = [[0, mine, 0]] 3566 for i in range(bins): 3567 lns2.append([-fs[i], edges[i], 0]) 3568 lns2.append([-fs[i], edges[i + 1], 0]) 3569 lns2.append([0, maxe, 0]) 3570 3571 if outline: 3572 rs.append(shapes.Line(lns1, c=lc, alpha=alpha, lw=lw).x(x)) 3573 rs.append(shapes.Line(lns2, c=lc, alpha=alpha, lw=lw).x(x)) 3574 3575 if fill: 3576 for i in range(bins): 3577 p0 = (-fs[i], edges[i], 0) 3578 p1 = (fs[i], edges[i + 1], 0) 3579 r = shapes.Rectangle(p0, p1).x(p0[0] + x) 3580 r.color(c).alpha(alpha).lighting("off") 3581 rs.append(r) 3582 3583 if centerline: 3584 cl = shapes.Line([0, mine, 0.01], [0, maxe, 0.01], c=lc, alpha=alpha, lw=2).x(x) 3585 rs.append(cl) 3586 3587 asse = Assembly(rs) 3588 asse.name = "Violin" 3589 return asse 3590 3591 3592def whisker(data, s=0.25, c="k", lw=2, bc="blue", alpha=0.25, r=5, jitter=True, horizontal=False): 3593 """ 3594 Generate a "whisker" bar from a 1-dimensional dataset. 3595 3596 Arguments: 3597 s : (float) 3598 size of the box 3599 c : (color) 3600 color of the lines 3601 lw : (float) 3602 line width 3603 bc : (color) 3604 color of the box 3605 alpha : (float) 3606 transparency of the box 3607 r : (float) 3608 point radius in pixels (use value 0 to disable) 3609 jitter : (bool) 3610 add some randomness to points to avoid overlap 3611 horizontal : (bool) 3612 set horizontal layout 3613 3614 Examples: 3615 - [whiskers.py](https://github.com/marcomusy/vedo/tree/master/examples/examples/pyplot/whiskers.py) 3616 3617  3618 """ 3619 xvals = np.zeros_like(np.asarray(data)) 3620 if jitter: 3621 xjit = np.random.randn(len(xvals)) * s / 9 3622 xjit = np.clip(xjit, -s / 2.1, s / 2.1) 3623 xvals += xjit 3624 3625 dmean = np.mean(data) 3626 dq05 = np.quantile(data, 0.05) 3627 dq25 = np.quantile(data, 0.25) 3628 dq75 = np.quantile(data, 0.75) 3629 dq95 = np.quantile(data, 0.95) 3630 3631 pts = None 3632 if r: 3633 pts = shapes.Points([xvals, data], c=c, r=r) 3634 3635 rec = shapes.Rectangle([-s / 2, dq25], [s / 2, dq75], c=bc, alpha=alpha) 3636 rec.GetProperty().LightingOff() 3637 rl = shapes.Line([[-s / 2, dq25], [s / 2, dq25], [s / 2, dq75], [-s / 2, dq75]], closed=True) 3638 l1 = shapes.Line([0, dq05, 0], [0, dq25, 0], c=c, lw=lw) 3639 l2 = shapes.Line([0, dq75, 0], [0, dq95, 0], c=c, lw=lw) 3640 lm = shapes.Line([-s / 2, dmean], [s / 2, dmean]) 3641 lns = merge(l1, l2, lm, rl) 3642 asse = Assembly([lns, rec, pts]) 3643 if horizontal: 3644 asse.rotate_z(-90) 3645 asse.name = "Whisker" 3646 asse.info["mean"] = dmean 3647 asse.info["quantile_05"] = dq05 3648 asse.info["quantile_25"] = dq25 3649 asse.info["quantile_75"] = dq75 3650 asse.info["quantile_95"] = dq95 3651 return asse 3652 3653 3654def streamplot( 3655 X, Y, U, V, direction="both", max_propagation=None, mode=1, lw=0.001, c=None, probes=() 3656): 3657 """ 3658 Generate a streamline plot of a vectorial field (U,V) defined at positions (X,Y). 3659 Returns a `Mesh` object. 3660 3661 Arguments: 3662 direction : (str) 3663 either "forward", "backward" or "both" 3664 max_propagation : (float) 3665 maximum physical length of the streamline 3666 lw : (float) 3667 line width in absolute units 3668 mode : (int) 3669 mode of varying the line width: 3670 - 0 - do not vary line width 3671 - 1 - vary line width by first vector component 3672 - 2 - vary line width vector magnitude 3673 - 3 - vary line width by absolute value of first vector component 3674 3675 Examples: 3676 - [plot_stream.py](https://github.com/marcomusy/vedo/tree/master/examples/examples/pyplot/plot_stream.py) 3677 3678  3679 """ 3680 n = len(X) 3681 m = len(Y[0]) 3682 if n != m: 3683 print("Limitation in streamplot(): only square grids are allowed.", n, m) 3684 raise RuntimeError() 3685 3686 xmin, xmax = X[0][0], X[-1][-1] 3687 ymin, ymax = Y[0][0], Y[-1][-1] 3688 3689 field = np.sqrt(U * U + V * V) 3690 3691 vol = vedo.Volume(field, dims=(n, n, 1)) 3692 3693 uf = np.ravel(U, order="F") 3694 vf = np.ravel(V, order="F") 3695 vects = np.c_[uf, vf, np.zeros_like(uf)] 3696 vol.pointdata["StreamPlotField"] = vects 3697 3698 if len(probes) == 0: 3699 probe = shapes.Grid(pos=((n - 1) / 2, (n - 1) / 2, 0), s=(n - 1, n - 1), res=(n - 1, n - 1)) 3700 else: 3701 if isinstance(probes, vedo.Points): 3702 probes = probes.points() 3703 else: 3704 probes = np.array(probes, dtype=float) 3705 if len(probes[0]) == 2: 3706 probes = np.c_[probes[:, 0], probes[:, 1], np.zeros(len(probes))] 3707 sv = [(n - 1) / (xmax - xmin), (n - 1) / (ymax - ymin), 1] 3708 probes = probes - [xmin, ymin, 0] 3709 probes = np.multiply(probes, sv) 3710 probe = vedo.Points(probes) 3711 3712 stream = vedo.shapes.StreamLines( 3713 vol, 3714 probe, 3715 tubes={"radius": lw, "mode": mode}, 3716 lw=lw, 3717 max_propagation=max_propagation, 3718 direction=direction, 3719 ) 3720 if c is not None: 3721 stream.color(c) 3722 else: 3723 stream.add_scalarbar() 3724 stream.lighting("off") 3725 3726 stream.scale([1 / (n - 1) * (xmax - xmin), 1 / (n - 1) * (ymax - ymin), 1]) 3727 stream.shift(xmin, ymin) 3728 return stream 3729 3730 3731def matrix( 3732 M, 3733 title="Matrix", 3734 xtitle="", 3735 ytitle="", 3736 xlabels=(), 3737 ylabels=(), 3738 xrotation=0, 3739 cmap="Reds", 3740 vmin=None, 3741 vmax=None, 3742 precision=2, 3743 font="Theemim", 3744 scale=0, 3745 scalarbar=True, 3746 lc="white", 3747 lw=0, 3748 c="black", 3749 alpha=1, 3750): 3751 """ 3752 Generate a matrix, or a 2D color-coded plot with bin labels. 3753 3754 Returns an `Assembly` object. 3755 3756 Arguments: 3757 M : (list, numpy array) 3758 the input array to visualize 3759 title : (str) 3760 title of the plot 3761 xtitle : (str) 3762 title of the horizontal colmuns 3763 ytitle : (str) 3764 title of the vertical rows 3765 xlabels : (list) 3766 individual string labels for each column. Must be of length m 3767 ylabels : (list) 3768 individual string labels for each row. Must be of length n 3769 xrotation : (float) 3770 rotation of the horizontal labels 3771 cmap : (str) 3772 color map name 3773 vmin : (float) 3774 minimum value of the colormap range 3775 vmax : (float) 3776 maximum value of the colormap range 3777 precision : (int) 3778 number of digits for the matrix entries or bins 3779 font : (str) 3780 font name. Check [available fonts here](https://vedo.embl.es/fonts). 3781 3782 scale : (float) 3783 size of the numeric entries or bin values 3784 scalarbar : (bool) 3785 add a scalar bar to the right of the plot 3786 lc : (str) 3787 color of the line separating the bins 3788 lw : (float) 3789 Width of the line separating the bins 3790 c : (str) 3791 text color 3792 alpha : (float) 3793 plot transparency 3794 3795 Examples: 3796 - [np_matrix.py](https://github.com/marcomusy/vedo/tree/master/examples/examples/pyplot/np_matrix.py) 3797 3798 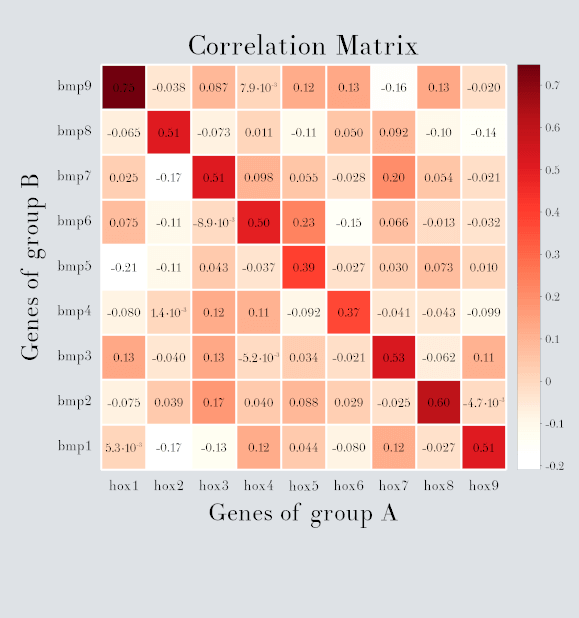 3799 """ 3800 M = np.asarray(M) 3801 n, m = M.shape 3802 gr = shapes.Grid(res=(m, n), s=(m / (m + n) * 2, n / (m + n) * 2), c=c, alpha=alpha) 3803 gr.wireframe(False).lc(lc).lw(lw) 3804 3805 matr = np.flip(np.flip(M), axis=1).ravel(order="C") 3806 gr.cmap(cmap, matr, on="cells", vmin=vmin, vmax=vmax) 3807 sbar = None 3808 if scalarbar: 3809 gr.add_scalarbar3d(title_font=font, label_font=font) 3810 sbar = gr.scalarbar 3811 labs = None 3812 if scale != 0: 3813 labs = gr.labels( 3814 on="cells", 3815 scale=scale / max(m, n), 3816 precision=precision, 3817 font=font, 3818 justify="center", 3819 c=c, 3820 ) 3821 labs.z(0.001) 3822 t = None 3823 if title: 3824 if title == "Matrix": 3825 title += " " + str(n) + "x" + str(m) 3826 t = shapes.Text3D(title, font=font, s=0.04, justify="bottom-center", c=c) 3827 t.shift(0, n / (m + n) * 1.05) 3828 3829 xlabs = None 3830 if len(xlabels) == m: 3831 xlabs = [] 3832 jus = "top-center" 3833 if xrotation > 44: 3834 jus = "right-center" 3835 for i in range(m): 3836 xl = shapes.Text3D(xlabels[i], font=font, s=0.02, justify=jus, c=c).rotate_z(xrotation) 3837 xl.shift((2 * i - m + 1) / (m + n), -n / (m + n) * 1.05) 3838 xlabs.append(xl) 3839 3840 ylabs = None 3841 if len(ylabels) == n: 3842 ylabels = list(reversed(ylabels)) 3843 ylabs = [] 3844 for i in range(n): 3845 yl = shapes.Text3D(ylabels[i], font=font, s=0.02, justify="right-center", c=c) 3846 yl.shift(-m / (m + n) * 1.05, (2 * i - n + 1) / (m + n)) 3847 ylabs.append(yl) 3848 3849 xt = None 3850 if xtitle: 3851 xt = shapes.Text3D(xtitle, font=font, s=0.035, justify="top-center", c=c) 3852 xt.shift(0, -n / (m + n) * 1.05) 3853 if xlabs is not None: 3854 y0, y1 = xlabs[0].ybounds() 3855 xt.shift(0, -(y1 - y0) - 0.55 / (m + n)) 3856 yt = None 3857 if ytitle: 3858 yt = shapes.Text3D(ytitle, font=font, s=0.035, justify="bottom-center", c=c).rotate_z(90) 3859 yt.shift(-m / (m + n) * 1.05, 0) 3860 if ylabs is not None: 3861 x0, x1 = ylabs[0].xbounds() 3862 yt.shift(-(x1 - x0) - 0.55 / (m + n), 0) 3863 asse = Assembly(gr, sbar, labs, t, xt, yt, xlabs, ylabs) 3864 asse.name = "Matrix" 3865 return asse 3866 3867 3868def CornerPlot(points, pos=1, s=0.2, title="", c="b", bg="k", lines=True, dots=True): 3869 """ 3870 Return a `vtkXYPlotActor` that is a plot of `x` versus `y`, 3871 where `points` is a list of `(x,y)` points. 3872 3873 Assign position following this convention: 3874 3875 - 1, topleft, 3876 - 2, topright, 3877 - 3, bottomleft, 3878 - 4, bottomright. 3879 """ 3880 if len(points) == 2: # passing [allx, ally] 3881 points = np.stack((points[0], points[1]), axis=1) 3882 3883 c = colors.get_color(c) # allow different codings 3884 array_x = vtk.vtkFloatArray() 3885 array_y = vtk.vtkFloatArray() 3886 array_x.SetNumberOfTuples(len(points)) 3887 array_y.SetNumberOfTuples(len(points)) 3888 for i, p in enumerate(points): 3889 array_x.InsertValue(i, p[0]) 3890 array_y.InsertValue(i, p[1]) 3891 field = vtk.vtkFieldData() 3892 field.AddArray(array_x) 3893 field.AddArray(array_y) 3894 data = vtk.vtkDataObject() 3895 data.SetFieldData(field) 3896 3897 xyplot = vtk.vtkXYPlotActor() 3898 xyplot.AddDataObjectInput(data) 3899 xyplot.SetDataObjectXComponent(0, 0) 3900 xyplot.SetDataObjectYComponent(0, 1) 3901 xyplot.SetXValuesToValue() 3902 xyplot.SetAdjustXLabels(0) 3903 xyplot.SetAdjustYLabels(0) 3904 xyplot.SetNumberOfXLabels(3) 3905 3906 xyplot.GetProperty().SetPointSize(5) 3907 xyplot.GetProperty().SetLineWidth(2) 3908 xyplot.GetProperty().SetColor(colors.get_color(bg)) 3909 xyplot.SetPlotColor(0, c[0], c[1], c[2]) 3910 3911 xyplot.SetXTitle(title) 3912 xyplot.SetYTitle("") 3913 xyplot.ExchangeAxesOff() 3914 xyplot.SetPlotPoints(dots) 3915 3916 if not lines: 3917 xyplot.PlotLinesOff() 3918 3919 if isinstance(pos, str): 3920 spos = 2 3921 if "top" in pos: 3922 if "left" in pos: 3923 spos = 1 3924 elif "right" in pos: 3925 spos = 2 3926 elif "bottom" in pos: 3927 if "left" in pos: 3928 spos = 3 3929 elif "right" in pos: 3930 spos = 4 3931 pos = spos 3932 if pos == 1: 3933 xyplot.GetPositionCoordinate().SetValue(0.0, 0.8, 0) 3934 elif pos == 2: 3935 xyplot.GetPositionCoordinate().SetValue(0.76, 0.8, 0) 3936 elif pos == 3: 3937 xyplot.GetPositionCoordinate().SetValue(0.0, 0.0, 0) 3938 elif pos == 4: 3939 xyplot.GetPositionCoordinate().SetValue(0.76, 0.0, 0) 3940 else: 3941 xyplot.GetPositionCoordinate().SetValue(pos[0], pos[1], 0) 3942 3943 xyplot.GetPosition2Coordinate().SetValue(s, s, 0) 3944 return xyplot 3945 3946 3947def CornerHistogram( 3948 values, 3949 bins=20, 3950 vrange=None, 3951 minbin=0, 3952 logscale=False, 3953 title="", 3954 c="g", 3955 bg="k", 3956 alpha=1, 3957 pos="bottom-left", 3958 s=0.175, 3959 lines=True, 3960 dots=False, 3961 nmax=None, 3962): 3963 """ 3964 Build a histogram from a list of values in n bins. 3965 The resulting object is a 2D actor. 3966 3967 Use `vrange` to restrict the range of the histogram. 3968 3969 Use `nmax` to limit the sampling to this max nr of entries 3970 3971 Use `pos` to assign its position: 3972 - 1, topleft, 3973 - 2, topright, 3974 - 3, bottomleft, 3975 - 4, bottomright, 3976 - (x, y), as fraction of the rendering window 3977 """ 3978 if hasattr(values, "inputdata"): 3979 values = utils.vtk2numpy(values.inputdata().GetPointData().GetScalars()) 3980 3981 n = values.shape[0] 3982 if nmax and nmax < n: 3983 # subsample: 3984 idxs = np.linspace(0, n, num=int(nmax), endpoint=False).astype(int) 3985 values = values[idxs] 3986 3987 fs, edges = np.histogram(values, bins=bins, range=vrange) 3988 3989 if minbin: 3990 fs = fs[minbin:-1] 3991 if logscale: 3992 fs = np.log10(fs + 1) 3993 pts = [] 3994 for i in range(len(fs)): 3995 pts.append([(edges[i] + edges[i + 1]) / 2, fs[i]]) 3996 3997 cplot = CornerPlot(pts, pos, s, title, c, bg, lines, dots) 3998 cplot.SetNumberOfYLabels(2) 3999 cplot.SetNumberOfXLabels(3) 4000 tprop = vtk.vtkTextProperty() 4001 tprop.SetColor(colors.get_color(bg)) 4002 tprop.SetFontFamily(vtk.VTK_FONT_FILE) 4003 tprop.SetFontFile(utils.get_font_path(vedo.settings.default_font)) 4004 tprop.SetOpacity(alpha) 4005 cplot.SetAxisTitleTextProperty(tprop) 4006 cplot.GetProperty().SetOpacity(alpha) 4007 cplot.GetXAxisActor2D().SetLabelTextProperty(tprop) 4008 cplot.GetXAxisActor2D().SetTitleTextProperty(tprop) 4009 cplot.GetXAxisActor2D().SetFontFactor(0.55) 4010 cplot.GetYAxisActor2D().SetLabelFactor(0.0) 4011 cplot.GetYAxisActor2D().LabelVisibilityOff() 4012 return cplot 4013 4014 4015class DirectedGraph(Assembly): 4016 """ 4017 Support for Directed Graphs. 4018 """ 4019 4020 def __init__(self, **kargs): 4021 """ 4022 A graph consists of a collection of nodes (without postional information) 4023 and a collection of edges connecting pairs of nodes. 4024 The task is to determine the node positions only based on their connections. 4025 4026 This class is derived from class `Assembly`, and it assembles 4 Mesh objects 4027 representing the graph, the node labels, edge labels and edge arrows. 4028 4029 Arguments: 4030 c : (color) 4031 Color of the Graph 4032 n : (int) 4033 number of the initial set of nodes 4034 layout : (int, str) 4035 layout in 4036 `['2d', 'fast2d', 'clustering2d', 'circular', 'circular3d', 'cone', 'force', 'tree']`. 4037 Each of these layouts has different available options. 4038 4039 --------------------------------------------------------------- 4040 .. note:: Options for layouts '2d', 'fast2d' and 'clustering2d' 4041 4042 Arguments: 4043 seed : (int) 4044 seed of the random number generator used to jitter point positions 4045 rest_distance : (float) 4046 manually set the resting distance 4047 nmax : (int) 4048 the maximum number of iterations to be used 4049 zrange : (list) 4050 expand 2d graph along z axis. 4051 4052 --------------------------------------------------------------- 4053 .. note:: Options for layouts 'circular', and 'circular3d': 4054 4055 Arguments: 4056 radius : (float) 4057 set the radius of the circles 4058 height : (float) 4059 set the vertical (local z) distance between the circles 4060 zrange : (float) 4061 expand 2d graph along z axis 4062 4063 --------------------------------------------------------------- 4064 .. note:: Options for layout 'cone' 4065 4066 Arguments: 4067 compactness : (float) 4068 ratio between the average width of a cone in the tree, 4069 and the height of the cone. 4070 compression : (bool) 4071 put children closer together, possibly allowing sub-trees to overlap. 4072 This is useful if the tree is actually the spanning tree of a graph. 4073 spacing : (float) 4074 space between layers of the tree 4075 4076 --------------------------------------------------------------- 4077 .. note:: Options for layout 'force' 4078 4079 Arguments: 4080 seed : (int) 4081 seed the random number generator used to jitter point positions 4082 bounds : (list) 4083 set the region in space in which to place the final graph 4084 nmax : (int) 4085 the maximum number of iterations to be used 4086 three_dimensional : (bool) 4087 allow optimization in the 3rd dimension too 4088 random_initial_points : (bool) 4089 use random positions within the graph bounds as initial points 4090 4091 Examples: 4092 - [lineage_graph.py](https://github.com/marcomusy/vedo/tree/master/examples/examples/pyplot/lineage_graph.py) 4093 4094  4095 4096 - [graph_network.py](https://github.com/marcomusy/vedo/tree/master/examples/examples/pyplot/graph_network.py) 4097 4098  4099 """ 4100 4101 vedo.Assembly.__init__(self) 4102 4103 self.nodes = [] 4104 self.edges = [] 4105 4106 self._node_labels = [] # holds strings 4107 self._edge_labels = [] 4108 self.edge_orientations = [] 4109 self.edge_glyph_position = 0.6 4110 4111 self.zrange = 0.0 4112 4113 self.rotX = 0 4114 self.rotY = 0 4115 self.rotZ = 0 4116 4117 self.arrow_scale = 0.15 4118 self.node_label_scale = None 4119 self.node_label_justify = "bottom-left" 4120 4121 self.edge_label_scale = None 4122 4123 self.mdg = vtk.vtkMutableDirectedGraph() 4124 4125 n = kargs.pop("n", 0) 4126 for _ in range(n): 4127 self.add_node() 4128 4129 self._c = kargs.pop("c", (0.3, 0.3, 0.3)) 4130 4131 self.gl = vtk.vtkGraphLayout() 4132 4133 self.font = kargs.pop("font", "") 4134 4135 s = kargs.pop("layout", "2d") 4136 if isinstance(s, int): 4137 ss = ["2d", "fast2d", "clustering2d", "circular", "circular3d", "cone", "force", "tree"] 4138 s = ss[s] 4139 self.layout = s 4140 4141 if "2d" in s: 4142 if "clustering" in s: 4143 self.strategy = vtk.vtkClustering2DLayoutStrategy() 4144 elif "fast" in s: 4145 self.strategy = vtk.vtkFast2DLayoutStrategy() 4146 else: 4147 self.strategy = vtk.vtkSimple2DLayoutStrategy() 4148 self.rotX = 180 4149 opt = kargs.pop("rest_distance", None) 4150 if opt is not None: 4151 self.strategy.SetRestDistance(opt) 4152 opt = kargs.pop("seed", None) 4153 if opt is not None: 4154 self.strategy.SetRandomSeed(opt) 4155 opt = kargs.pop("nmax", None) 4156 if opt is not None: 4157 self.strategy.SetMaxNumberOfIterations(opt) 4158 self.zrange = kargs.pop("zrange", 0) 4159 4160 elif "circ" in s: 4161 if "3d" in s: 4162 self.strategy = vtk.vtkSimple3DCirclesStrategy() 4163 self.strategy.SetDirection(0, 0, -1) 4164 self.strategy.SetAutoHeight(True) 4165 self.strategy.SetMethod(1) 4166 self.rotX = -90 4167 opt = kargs.pop("radius", None) # float 4168 if opt is not None: 4169 self.strategy.SetMethod(0) 4170 self.strategy.SetRadius(opt) # float 4171 opt = kargs.pop("height", None) 4172 if opt is not None: 4173 self.strategy.SetAutoHeight(False) 4174 self.strategy.SetHeight(opt) # float 4175 else: 4176 self.strategy = vtk.vtkCircularLayoutStrategy() 4177 self.zrange = kargs.pop("zrange", 0) 4178 4179 elif "cone" in s: 4180 self.strategy = vtk.vtkConeLayoutStrategy() 4181 self.rotX = 180 4182 opt = kargs.pop("compactness", None) 4183 if opt is not None: 4184 self.strategy.SetCompactness(opt) 4185 opt = kargs.pop("compression", None) 4186 if opt is not None: 4187 self.strategy.SetCompression(opt) 4188 opt = kargs.pop("spacing", None) 4189 if opt is not None: 4190 self.strategy.SetSpacing(opt) 4191 4192 elif "force" in s: 4193 self.strategy = vtk.vtkForceDirectedLayoutStrategy() 4194 opt = kargs.pop("seed", None) 4195 if opt is not None: 4196 self.strategy.SetRandomSeed(opt) 4197 opt = kargs.pop("bounds", None) 4198 if opt is not None: 4199 self.strategy.SetAutomaticBoundsComputation(False) 4200 self.strategy.SetGraphBounds(opt) # list 4201 opt = kargs.pop("nmax", None) 4202 if opt is not None: 4203 self.strategy.SetMaxNumberOfIterations(opt) # int 4204 opt = kargs.pop("three_dimensional", True) 4205 if opt is not None: 4206 self.strategy.SetThreeDimensionalLayout(opt) # bool 4207 opt = kargs.pop("random_initial_points", None) 4208 if opt is not None: 4209 self.strategy.SetRandomInitialPoints(opt) # bool 4210 4211 elif "tree" in s: 4212 self.strategy = vtk.vtkSpanTreeLayoutStrategy() 4213 self.rotX = 180 4214 4215 else: 4216 vedo.logger.error(f"Cannot understand layout {s}. Available layouts:") 4217 vedo.logger.error("[2d,fast2d,clustering2d,circular,circular3d,cone,force,tree]") 4218 raise RuntimeError() 4219 4220 self.gl.SetLayoutStrategy(self.strategy) 4221 4222 if len(kargs) > 0: 4223 vedo.logger.error(f"Cannot understand options: {kargs}") 4224 4225 def add_node(self, label="id"): 4226 """Add a new node to the `Graph`.""" 4227 v = self.mdg.AddVertex() # vtk calls it vertex.. 4228 self.nodes.append(v) 4229 if label == "id": 4230 label = int(v) 4231 self._node_labels.append(str(label)) 4232 return v 4233 4234 def add_edge(self, v1, v2, label=""): 4235 """Add a new edge between to nodes. 4236 An extra node is created automatically if needed.""" 4237 nv = len(self.nodes) 4238 if v1 >= nv: 4239 for _ in range(nv, v1 + 1): 4240 self.add_node() 4241 nv = len(self.nodes) 4242 if v2 >= nv: 4243 for _ in range(nv, v2 + 1): 4244 self.add_node() 4245 e = self.mdg.AddEdge(v1, v2) 4246 self.edges.append(e) 4247 self._edge_labels.append(str(label)) 4248 return e 4249 4250 def add_child(self, v, node_label="id", edge_label=""): 4251 """Add a new edge to a new node as its child. 4252 The extra node is created automatically if needed.""" 4253 nv = len(self.nodes) 4254 if v >= nv: 4255 for _ in range(nv, v + 1): 4256 self.add_node() 4257 child = self.mdg.AddChild(v) 4258 self.edges.append((v, child)) 4259 self.nodes.append(child) 4260 if node_label == "id": 4261 node_label = int(child) 4262 self._node_labels.append(str(node_label)) 4263 self._edge_labels.append(str(edge_label)) 4264 return child 4265 4266 def build(self): 4267 """ 4268 Build the `DirectedGraph(Assembly)`. 4269 Accessory objects are also created for labels and arrows. 4270 """ 4271 self.gl.SetZRange(self.zrange) 4272 self.gl.SetInputData(self.mdg) 4273 self.gl.Update() 4274 4275 gr2poly = vtk.vtkGraphToPolyData() 4276 gr2poly.EdgeGlyphOutputOn() 4277 gr2poly.SetEdgeGlyphPosition(self.edge_glyph_position) 4278 gr2poly.SetInputData(self.gl.GetOutput()) 4279 gr2poly.Update() 4280 4281 dgraph = Mesh(gr2poly.GetOutput(0)) 4282 # dgraph.clean() # WRONG!!! dont uncomment 4283 dgraph.flat().color(self._c).lw(2) 4284 dgraph.name = "DirectedGraph" 4285 4286 diagsz = self.diagonal_size() / 1.42 4287 if not diagsz: 4288 return None 4289 4290 dgraph.SetScale(1 / diagsz) 4291 if self.rotX: 4292 dgraph.rotate_x(self.rotX) 4293 if self.rotY: 4294 dgraph.rotate_y(self.rotY) 4295 if self.rotZ: 4296 dgraph.rotate_z(self.rotZ) 4297 4298 vecs = gr2poly.GetOutput(1).GetPointData().GetVectors() 4299 self.edge_orientations = utils.vtk2numpy(vecs) 4300 4301 # Use Glyph3D to repeat the glyph on all edges. 4302 arrows = None 4303 if self.arrow_scale: 4304 arrow_source = vtk.vtkGlyphSource2D() 4305 arrow_source.SetGlyphTypeToEdgeArrow() 4306 arrow_source.SetScale(self.arrow_scale) 4307 arrow_source.Update() 4308 arrow_glyph = vtk.vtkGlyph3D() 4309 arrow_glyph.SetInputData(0, gr2poly.GetOutput(1)) 4310 arrow_glyph.SetInputData(1, arrow_source.GetOutput()) 4311 arrow_glyph.Update() 4312 arrows = Mesh(arrow_glyph.GetOutput()) 4313 arrows.SetScale(1 / diagsz) 4314 arrows.lighting("off").color(self._c) 4315 if self.rotX: 4316 arrows.rotate_x(self.rotX) 4317 if self.rotY: 4318 arrows.rotate_y(self.rotY) 4319 if self.rotZ: 4320 arrows.rotate_z(self.rotZ) 4321 arrows.name = "DirectedGraphArrows" 4322 4323 node_labels = dgraph.labels( 4324 self._node_labels, 4325 scale=self.node_label_scale, 4326 precision=0, 4327 font=self.font, 4328 justify=self.node_label_justify, 4329 ) 4330 node_labels.color(self._c).pickable(True) 4331 node_labels.name = "DirectedGraphNodeLabels" 4332 4333 edge_labels = dgraph.labels( 4334 self._edge_labels, on="cells", scale=self.edge_label_scale, precision=0, font=self.font 4335 ) 4336 edge_labels.color(self._c).pickable(True) 4337 edge_labels.name = "DirectedGraphEdgeLabels" 4338 4339 Assembly.__init__(self, [dgraph, node_labels, edge_labels, arrows]) 4340 self.name = "DirectedGraphAssembly" 4341 return self
96class Figure(Assembly): 97 """Format class for figures.""" 98 99 def __init__(self, xlim, ylim, aspect=4 / 3, padding=(0.05, 0.05, 0.05, 0.05), **kwargs): 100 """ 101 Create an empty formatted figure for plotting. 102 103 Arguments: 104 xlim : (list) 105 range of the x-axis as [x0, x1] 106 ylim : (list) 107 range of the y-axis as [y0, y1] 108 aspect : (float, str) 109 the desired aspect ratio of the histogram. Default is 4/3. 110 Use `aspect="equal"` to force the same units in x and y. 111 padding : (float, list) 112 keep a padding space from the axes (as a fraction of the axis size). 113 This can be a list of four numbers. 114 xtitle : (str) 115 title for the x-axis, can also be set using `axes=dict(xtitle="my x axis")` 116 ytitle : (str) 117 title for the y-axis, can also be set using `axes=dict(ytitle="my y axis")` 118 grid : (bool) 119 show the background grid for the axes, can also be set using `axes=dict(xygrid=True)` 120 axes : (dict) 121 an extra dictionary of options for the `vedo.addons.Axes` object 122 """ 123 124 self.verbose = True # printing to stdout on every mouse click 125 126 self.xlim = np.asarray(xlim) 127 self.ylim = np.asarray(ylim) 128 self.aspect = aspect 129 self.padding = padding 130 if not utils.is_sequence(self.padding): 131 self.padding = [self.padding, self.padding, self.padding, self.padding] 132 133 self.force_scaling_types = ( 134 shapes.Glyph, 135 shapes.Line, 136 shapes.Rectangle, 137 shapes.DashedLine, 138 shapes.Tube, 139 shapes.Ribbon, 140 shapes.GeoCircle, 141 shapes.Arc, 142 shapes.Grid, 143 # shapes.Arrows, # todo 144 # shapes.Arrows2D, # todo 145 shapes.Brace, # todo 146 ) 147 148 options = dict(kwargs) 149 150 self.title = options.pop("title", "") 151 self.xtitle = options.pop("xtitle", " ") 152 self.ytitle = options.pop("ytitle", " ") 153 number_of_divisions = 6 154 155 self.legend = None 156 self.labels = [] 157 self.label = options.pop("label", None) 158 if self.label: 159 self.labels = [self.label] 160 161 self.axopts = options.pop("axes", {}) 162 if isinstance(self.axopts, (bool, int, float)): 163 if self.axopts: 164 self.axopts = {} 165 if self.axopts or isinstance(self.axopts, dict): 166 number_of_divisions = self.axopts.pop("number_of_divisions", number_of_divisions) 167 168 self.axopts["xtitle"] = self.xtitle 169 self.axopts["ytitle"] = self.ytitle 170 171 if "xygrid" not in self.axopts: ## modify the default 172 self.axopts["xygrid"] = options.pop("grid", False) 173 174 if "xygrid_transparent" not in self.axopts: ## modify the default 175 self.axopts["xygrid_transparent"] = True 176 177 if "xtitle_position" not in self.axopts: ## modify the default 178 self.axopts["xtitle_position"] = 0.5 179 self.axopts["xtitle_justify"] = "top-center" 180 181 if "ytitle_position" not in self.axopts: ## modify the default 182 self.axopts["ytitle_position"] = 0.5 183 self.axopts["ytitle_justify"] = "bottom-center" 184 185 if self.label: 186 if "c" in self.axopts: 187 self.label.tcolor = self.axopts["c"] 188 189 x0, x1 = self.xlim 190 y0, y1 = self.ylim 191 dx = x1 - x0 192 dy = y1 - y0 193 x0lim, x1lim = (x0 - self.padding[0] * dx, x1 + self.padding[1] * dx) 194 y0lim, y1lim = (y0 - self.padding[2] * dy, y1 + self.padding[3] * dy) 195 dy = y1lim - y0lim 196 197 self.axes = None 198 if xlim[0] >= xlim[1] or ylim[0] >= ylim[1]: 199 vedo.logger.warning(f"Null range for Figure {self.title}... returning an empty Assembly.") 200 Assembly.__init__(self) 201 self.yscale = 0 202 return 203 204 if aspect == "equal": 205 self.aspect = dx / dy # so that yscale becomes 1 206 207 self.yscale = dx / dy / self.aspect 208 209 y0lim *= self.yscale 210 y1lim *= self.yscale 211 212 self.x0lim = x0lim 213 self.x1lim = x1lim 214 self.y0lim = y0lim 215 self.y1lim = y1lim 216 217 self.ztolerance = options.pop("ztolerance", None) 218 if self.ztolerance is None: 219 self.ztolerance = dx / 5000 220 221 ############## create axes 222 if self.axopts: 223 axes_opts = self.axopts 224 if self.axopts is True or self.axopts == 1: 225 axes_opts = {} 226 227 tp, ts = utils.make_ticks(y0lim / self.yscale, y1lim / self.yscale, number_of_divisions) 228 labs = [] 229 for i in range(1, len(tp) - 1): 230 ynew = utils.lin_interpolate(tp[i], [0, 1], [y0lim, y1lim]) 231 labs.append([ynew, ts[i]]) 232 233 if self.title: 234 axes_opts["htitle"] = self.title 235 axes_opts["y_values_and_labels"] = labs 236 axes_opts["xrange"] = (x0lim, x1lim) 237 axes_opts["yrange"] = (y0lim, y1lim) 238 axes_opts["zrange"] = (0, 0) 239 axes_opts["y_use_bounds"] = True 240 241 if "c" not in axes_opts and "ac" in options: 242 axes_opts["c"] = options["ac"] 243 244 self.axes = addons.Axes(**axes_opts) 245 246 Assembly.__init__(self, [self.axes]) 247 self.name = "Figure" 248 249 vedo.last_figure = self if settings.remember_last_figure_format else None 250 return 251 252 def _repr_html_(self): 253 """ 254 HTML representation of the Figure object for Jupyter Notebooks. 255 256 Returns: 257 HTML text with the image and some properties. 258 """ 259 import io 260 import base64 261 from PIL import Image 262 263 library_name = "vedo.pyplot.Figure" 264 help_url = "https://vedo.embl.es/docs/vedo/pyplot.html#Figure" 265 266 arr = self.thumbnail(zoom=1.1) 267 268 im = Image.fromarray(arr) 269 buffered = io.BytesIO() 270 im.save(buffered, format="PNG", quality=100) 271 encoded = base64.b64encode(buffered.getvalue()).decode("utf-8") 272 url = "data:image/png;base64," + encoded 273 image = f"<img src='{url}'></img>" 274 275 bounds = "<br/>".join( 276 [ 277 vedo.utils.precision(min_x, 4) + " ... " + vedo.utils.precision(max_x, 4) 278 for min_x, max_x in zip(self.bounds()[::2], self.bounds()[1::2]) 279 ] 280 ) 281 282 help_text = "" 283 if self.name: 284 help_text += f"<b> {self.name}:   </b>" 285 help_text += '<b><a href="' + help_url + '" target="_blank">' + library_name + "</a></b>" 286 if self.filename: 287 dots = "" 288 if len(self.filename) > 30: 289 dots = "..." 290 help_text += f"<br/><code><i>({dots}{self.filename[-30:]})</i></code>" 291 292 all = [ 293 "<table>", 294 "<tr>", 295 "<td>", 296 image, 297 "</td>", 298 "<td style='text-align: center; vertical-align: center;'><br/>", 299 help_text, 300 "<table>", 301 "<tr><td><b> nr. of parts </b></td><td>" + str(self.GetNumberOfPaths()) + "</td></tr>", 302 "<tr><td><b> position </b></td><td>" + str(self.GetPosition()) + "</td></tr>", 303 "<tr><td><b> x-limits </b></td><td>" + utils.precision(self.xlim, 4) + "</td></tr>", 304 "<tr><td><b> y-limits </b></td><td>" + utils.precision(self.ylim, 4) + "</td></tr>", 305 "<tr><td><b> world bounds </b> <br/> (x/y/z) </td><td>" + str(bounds) + "</td></tr>", 306 "</table>", 307 "</table>", 308 ] 309 return "\n".join(all) 310 311 def __add__(self, *obj): 312 # just to avoid confusion, supersede Assembly.__add__ 313 return self.__iadd__(*obj) 314 315 def __iadd__(self, *obj): 316 if len(obj) == 1 and isinstance(obj[0], Figure): 317 return self._check_unpack_and_insert(obj[0]) 318 319 obj = utils.flatten(obj) 320 return self.insert(*obj) 321 322 def _check_unpack_and_insert(self, fig): 323 324 if fig.label: 325 self.labels.append(fig.label) 326 327 if abs(self.yscale - fig.yscale) > 0.0001: 328 329 colors.printc(":bomb:ERROR: adding incompatible Figure. Y-scales are different:", 330 c='r', invert=True) 331 colors.printc(" first figure:", self.yscale, c='r') 332 colors.printc(" second figure:", fig.yscale, c='r') 333 334 colors.printc("One or more of these parameters can be the cause:", c="r") 335 if list(self.xlim) != list(fig.xlim): 336 colors.printc("xlim --------------------------------------------\n", 337 " first figure:", self.xlim, "\n", 338 " second figure:", fig.xlim, c='r') 339 if list(self.ylim) != list(fig.ylim): 340 colors.printc("ylim --------------------------------------------\n", 341 " first figure:", self.ylim, "\n", 342 " second figure:", fig.ylim, c='r') 343 if list(self.padding) != list(fig.padding): 344 colors.printc("padding -----------------------------------------\n", 345 " first figure:", self.padding, 346 " second figure:", fig.padding, c='r') 347 if self.aspect != fig.aspect: 348 colors.printc("aspect ------------------------------------------\n", 349 " first figure:", self.aspect, "\n", 350 " second figure:", fig.aspect, c='r') 351 352 colors.printc("\n:idea: Consider using fig2 = histogram(..., like=fig1)", c="r") 353 colors.printc(" Or fig += histogram(..., like=fig)\n", c="r") 354 return self 355 356 offset = self.zbounds()[1] + self.ztolerance 357 358 for ele in fig.unpack(): 359 if "Axes" in ele.name: 360 continue 361 ele.z(offset) 362 self.insert(ele, rescale=False) 363 364 return self 365 366 def insert(self, *objs, rescale=True, as3d=True, adjusted=False, cut=True): 367 """ 368 Insert objects into a Figure. 369 370 The recommended syntax is to use "+=", which calls `insert()` under the hood. 371 If a whole Figure is added with "+=", it is unpacked and its objects are added 372 one by one. 373 374 Arguments: 375 rescale : (bool) 376 rescale the y axis position while inserting the object. 377 as3d : (bool) 378 if True keep the aspect ratio of the 3d object, otherwise stretch it in y. 379 adjusted : (bool) 380 adjust the scaling according to the shortest axis 381 cut : (bool) 382 cut off the parts of the object which go beyond the axes frame. 383 """ 384 for a in objs: 385 386 if a in self.actors: 387 # should not add twice the same object in plot 388 continue 389 390 if isinstance(a, vedo.Points): # hacky way to identify Points 391 if a.ncells == a.npoints: 392 poly = a.polydata(False) 393 if poly.GetNumberOfPolys() == 0 and poly.GetNumberOfLines() == 0: 394 as3d = False 395 rescale = True 396 397 if isinstance(a, (shapes.Arrow, shapes.Arrow2D)): 398 # discard input Arrow and substitute it with a brand new one 399 # (because scaling would fatally distort the shape) 400 prop = a.GetProperty() 401 prop.LightingOff() 402 py = a.base[1] 403 a.top[1] = (a.top[1] - py) * self.yscale + py 404 b = shapes.Arrow2D(a.base, a.top, s=a.s, fill=a.fill).z(a.z()) 405 b.SetProperty(prop) 406 b.y(py * self.yscale) 407 a = b 408 409 # elif isinstance(a, shapes.Rectangle) and a.radius is not None: 410 # # discard input Rectangle and substitute it with a brand new one 411 # # (because scaling would fatally distort the shape of the corners) 412 # py = a.corner1[1] 413 # rx1,ry1,rz1 = a.corner1 414 # rx2,ry2,rz2 = a.corner2 415 # ry2 = (ry2-py) * self.yscale + py 416 # b = shapes.Rectangle([rx1,0,rz1], [rx2,ry2,rz2], radius=a.radius).z(a.z()) 417 # b.SetProperty(a.GetProperty()) 418 # b.y(py / self.yscale) 419 # a = b 420 421 else: 422 423 if rescale: 424 425 if not isinstance(a, Figure): 426 427 if as3d and not isinstance(a, self.force_scaling_types): 428 if adjusted: 429 scl = np.min([1, self.yscale]) 430 else: 431 scl = self.yscale 432 433 a.scale(scl) 434 435 else: 436 a.scale([1, self.yscale, 1]) 437 438 # shift it in y 439 a.y(a.y() * self.yscale) 440 441 if cut: 442 try: 443 bx0, bx1, by0, by1, _, _ = a.bounds() 444 if self.y0lim > by0: 445 a.cut_with_plane([0, self.y0lim, 0], [0, 1, 0]) 446 if self.y1lim < by1: 447 a.cut_with_plane([0, self.y1lim, 0], [0, -1, 0]) 448 if self.x0lim > bx0: 449 a.cut_with_plane([self.x0lim, 0, 0], [1, 0, 0]) 450 if self.x1lim < bx1: 451 a.cut_with_plane([self.x1lim, 0, 0], [-1, 0, 0]) 452 except: 453 # print("insert(): cannot cut", [a]) 454 pass 455 456 self.AddPart(a) 457 self.actors.append(a) 458 459 return self 460 461 def add_label(self, text, c=None, marker="", mc="black"): 462 """ 463 Manually add en entry label to the legend. 464 465 Arguments: 466 text : (str) 467 text string for the label. 468 c : (str) 469 color of the text 470 marker : (str), Mesh 471 a marker char or a Mesh object to be used as marker 472 mc : (str) 473 color for the marker 474 """ 475 newlabel = LabelData() 476 newlabel.text = text.replace("\n", " ") 477 newlabel.tcolor = c 478 newlabel.marker = marker 479 newlabel.mcolor = mc 480 self.labels.append(newlabel) 481 return self 482 483 def add_legend( 484 self, 485 pos="top-right", 486 relative=True, 487 font=None, 488 s=1, 489 c=None, 490 vspace=1.75, 491 padding=0.1, 492 radius=0, 493 alpha=1, 494 bc="k7", 495 lw=1, 496 lc="k4", 497 z=0, 498 ): 499 """ 500 Add existing labels to form a legend box. 501 Labels have been previously filled with eg: `plot(..., label="text")` 502 503 Arguments: 504 pos : (str, list) 505 A string or 2D coordinates. The default is "top-right". 506 relative : (bool) 507 control whether `pos` is absolute or relative, e.i. normalized 508 to the x and y ranges so that x and y in `pos=[x,y]` should be 509 both in the range [0,1]. 510 This flag is ignored if a string despcriptor is passed. 511 Default is True. 512 font : (str, int) 513 font name or number. 514 Check [available fonts here](https://vedo.embl.es/fonts). 515 s : (float) 516 global size of the legend 517 c : (str) 518 color of the text 519 vspace : (float) 520 vertical spacing of lines 521 padding : (float) 522 padding of the box as a fraction of the text size 523 radius : (float) 524 border radius of the box 525 alpha : (float) 526 opacity of the box. Values below 1 may cause poor rendering 527 because of antialiasing. 528 Use alpha = 0 to remove the box. 529 bc : (str) 530 box color 531 lw : (int) 532 border line width of the box in pixel units 533 lc : (int) 534 border line color of the box 535 z : (float) 536 set the zorder as z position (useful to avoid overlap) 537 """ 538 sx = self.x1lim - self.x0lim 539 s = s * sx / 55 # so that input can be about 1 540 541 ds = 0 542 texts = [] 543 mks = [] 544 for i, t in enumerate(self.labels): 545 label = self.labels[i] 546 t = label.text 547 548 if label.tcolor is not None: 549 c = label.tcolor 550 551 tx = vedo.shapes.Text3D(t, s=s, c=c, justify="center-left", font=font) 552 y0, y1 = tx.ybounds() 553 ds = max(y1 - y0, ds) 554 texts.append(tx) 555 556 mk = label.marker 557 if isinstance(mk, vedo.Points): 558 mk = mk.clone(deep=False).lighting("off") 559 cm = mk.center_of_mass() 560 ty0, ty1 = tx.ybounds() 561 oby0, oby1 = mk.ybounds() 562 mk.shift(-cm) 563 mk.SetOrigin(cm) 564 mk.scale((ty1 - ty0) / (oby1 - oby0)) 565 mk.scale([1.1, 1.1, 0.01]) 566 elif mk == "-": 567 mk = vedo.shapes.Marker(mk, s=s * 2) 568 mk.color(label.mcolor) 569 else: 570 mk = vedo.shapes.Marker(mk, s=s) 571 mk.color(label.mcolor) 572 mks.append(mk) 573 574 for i, tx in enumerate(texts): 575 tx.shift(0, -(i + 0) * ds * vspace) 576 577 for i, mk in enumerate(mks): 578 mk.shift(-ds * 1.75, -(i + 0) * ds * vspace, 0) 579 580 acts = texts + mks 581 582 aleg = Assembly(acts) # .show(axes=1).close() 583 x0, x1, y0, y1, _, _ = aleg.GetBounds() 584 585 if alpha: 586 dx = x1 - x0 587 dy = y1 - y0 588 589 if not utils.is_sequence(padding): 590 padding = [padding] * 4 591 padding = min(padding) 592 padding = min(padding * dx, padding * dy) 593 if len(self.labels) == 1: 594 padding *= 4 595 x0 -= padding 596 x1 += padding 597 y0 -= padding 598 y1 += padding 599 600 box = shapes.Rectangle([x0, y0], [x1, y1], radius=radius, c=bc, alpha=alpha) 601 box.shift(0, 0, -dy / 100).pickable(False) 602 if lc: 603 box.lc(lc).lw(lw) 604 aleg.AddPart(box) 605 606 xlim = self.xlim 607 ylim = self.ylim 608 if isinstance(pos, str): 609 px, py = 0, 0 610 rx, ry = (xlim[1] + xlim[0]) / 2, (ylim[1] + ylim[0]) / 2 611 shx, shy = 0, 0 612 if "top" in pos: 613 if "cent" in pos: 614 px, py = rx, ylim[1] 615 shx, shy = (x0 + x1) / 2, y1 616 elif "left" in pos: 617 px, py = xlim[0], ylim[1] 618 shx, shy = x0, y1 619 else: # "right" 620 px, py = xlim[1], ylim[1] 621 shx, shy = x1, y1 622 elif "bot" in pos: 623 if "left" in pos: 624 px, py = xlim[0], ylim[0] 625 shx, shy = x0, y0 626 elif "right" in pos: 627 px, py = xlim[1], ylim[0] 628 shx, shy = x1, y0 629 else: # "cent" 630 px, py = rx, ylim[0] 631 shx, shy = (x0 + x1) / 2, y0 632 elif "cent" in pos: 633 if "left" in pos: 634 px, py = xlim[0], ry 635 shx, shy = x0, (y0 + y1) / 2 636 elif "right" in pos: 637 px, py = xlim[1], ry 638 shx, shy = x1, (y0 + y1) / 2 639 else: 640 vedo.logger.error(f"in add_legend(), cannot understand {pos}") 641 raise RuntimeError 642 643 else: 644 645 if relative: 646 rx, ry = pos[0], pos[1] 647 px = (xlim[1] - xlim[0]) * rx + xlim[0] 648 py = (ylim[1] - ylim[0]) * ry + ylim[0] 649 z *= xlim[1] - xlim[0] 650 else: 651 px, py = pos[0], pos[1] 652 shx, shy = x0, y1 653 654 aleg.pos(px - shx, py * self.yscale - shy, self.z() + sx / 50 + z) 655 656 self.insert(aleg, rescale=False, cut=False) 657 self.legend = aleg 658 aleg.name = "Legend" 659 return self 660 661 def as2d(self, pos="bottom-left", scale=1, padding=0.05): 662 """ 663 Convert the Figure into a 2D static object (a 2D Assembly). 664 665 Arguments: 666 pos : (str, list) 667 position in 2D, as atring or list (x,y). 668 Any combination of "center", "top", "bottom", "left" and "right" will work. 669 The center of the renderer is [0,0] while top-right is [1,1]. 670 scale : (float) 671 scaling factor 672 padding : (float, list) 673 a single value or a list (xpad, ypad) 674 675 Returns: 676 `Group` object. 677 """ 678 x0, x1 = self.xbounds() 679 y0, y1 = self.ybounds() 680 pp = self.pos() 681 x0 -= pp[0] 682 x1 -= pp[0] 683 y0 -= pp[1] 684 y1 -= pp[1] 685 686 if not utils.is_sequence(padding): 687 padding = (padding, padding) 688 padding = np.array(padding) 689 690 if "cent" in pos: 691 offset = [(x0 + x1) / 2, (y0 + y1) / 2] 692 position = [0, 0] 693 if "right" in pos: 694 offset[0] = x1 695 position = [1 - padding[0], 0] 696 if "left" in pos: 697 offset[0] = x0 698 position = [-1 + padding[0], 0] 699 if "top" in pos: 700 offset[1] = y1 701 position = [0, 1 - padding[1]] 702 if "bottom" in pos: 703 offset[1] = y0 704 position = [0, -1 + padding[1]] 705 elif "top" in pos: 706 if "right" in pos: 707 offset = [x1, y1] 708 position = [1, 1] - padding 709 elif "left" in pos: 710 offset = [x0, y1] 711 position = [-1 + padding[0], 1 - padding[1]] 712 else: 713 raise ValueError(f"incomplete position pos='{pos}'") 714 elif "bottom" in pos: 715 if "right" in pos: 716 offset = [x1, y0] 717 position = [1 - padding[0], -1 + padding[1]] 718 elif "left" in pos: 719 offset = [x0, y0] 720 position = [-1, -1] + padding 721 else: 722 raise ValueError(f"incomplete position pos='{pos}'") 723 else: 724 offset = [0, 0] 725 position = pos 726 727 scanned = [] 728 group = Group() 729 for a in self.recursive_unpack(): 730 if a in scanned: 731 continue 732 if not isinstance(a, vedo.Points): 733 continue 734 if a.npoints == 0: 735 continue 736 if a.GetProperty().GetRepresentation() == 1: 737 # wireframe is not rendered correctly in 2d 738 continue 739 a2d = _to2d(a, offset, scale * 550 / (x1 - x0)) 740 a2d.SetPosition(position) 741 group += a2d 742 return group
Format class for figures.
99 def __init__(self, xlim, ylim, aspect=4 / 3, padding=(0.05, 0.05, 0.05, 0.05), **kwargs): 100 """ 101 Create an empty formatted figure for plotting. 102 103 Arguments: 104 xlim : (list) 105 range of the x-axis as [x0, x1] 106 ylim : (list) 107 range of the y-axis as [y0, y1] 108 aspect : (float, str) 109 the desired aspect ratio of the histogram. Default is 4/3. 110 Use `aspect="equal"` to force the same units in x and y. 111 padding : (float, list) 112 keep a padding space from the axes (as a fraction of the axis size). 113 This can be a list of four numbers. 114 xtitle : (str) 115 title for the x-axis, can also be set using `axes=dict(xtitle="my x axis")` 116 ytitle : (str) 117 title for the y-axis, can also be set using `axes=dict(ytitle="my y axis")` 118 grid : (bool) 119 show the background grid for the axes, can also be set using `axes=dict(xygrid=True)` 120 axes : (dict) 121 an extra dictionary of options for the `vedo.addons.Axes` object 122 """ 123 124 self.verbose = True # printing to stdout on every mouse click 125 126 self.xlim = np.asarray(xlim) 127 self.ylim = np.asarray(ylim) 128 self.aspect = aspect 129 self.padding = padding 130 if not utils.is_sequence(self.padding): 131 self.padding = [self.padding, self.padding, self.padding, self.padding] 132 133 self.force_scaling_types = ( 134 shapes.Glyph, 135 shapes.Line, 136 shapes.Rectangle, 137 shapes.DashedLine, 138 shapes.Tube, 139 shapes.Ribbon, 140 shapes.GeoCircle, 141 shapes.Arc, 142 shapes.Grid, 143 # shapes.Arrows, # todo 144 # shapes.Arrows2D, # todo 145 shapes.Brace, # todo 146 ) 147 148 options = dict(kwargs) 149 150 self.title = options.pop("title", "") 151 self.xtitle = options.pop("xtitle", " ") 152 self.ytitle = options.pop("ytitle", " ") 153 number_of_divisions = 6 154 155 self.legend = None 156 self.labels = [] 157 self.label = options.pop("label", None) 158 if self.label: 159 self.labels = [self.label] 160 161 self.axopts = options.pop("axes", {}) 162 if isinstance(self.axopts, (bool, int, float)): 163 if self.axopts: 164 self.axopts = {} 165 if self.axopts or isinstance(self.axopts, dict): 166 number_of_divisions = self.axopts.pop("number_of_divisions", number_of_divisions) 167 168 self.axopts["xtitle"] = self.xtitle 169 self.axopts["ytitle"] = self.ytitle 170 171 if "xygrid" not in self.axopts: ## modify the default 172 self.axopts["xygrid"] = options.pop("grid", False) 173 174 if "xygrid_transparent" not in self.axopts: ## modify the default 175 self.axopts["xygrid_transparent"] = True 176 177 if "xtitle_position" not in self.axopts: ## modify the default 178 self.axopts["xtitle_position"] = 0.5 179 self.axopts["xtitle_justify"] = "top-center" 180 181 if "ytitle_position" not in self.axopts: ## modify the default 182 self.axopts["ytitle_position"] = 0.5 183 self.axopts["ytitle_justify"] = "bottom-center" 184 185 if self.label: 186 if "c" in self.axopts: 187 self.label.tcolor = self.axopts["c"] 188 189 x0, x1 = self.xlim 190 y0, y1 = self.ylim 191 dx = x1 - x0 192 dy = y1 - y0 193 x0lim, x1lim = (x0 - self.padding[0] * dx, x1 + self.padding[1] * dx) 194 y0lim, y1lim = (y0 - self.padding[2] * dy, y1 + self.padding[3] * dy) 195 dy = y1lim - y0lim 196 197 self.axes = None 198 if xlim[0] >= xlim[1] or ylim[0] >= ylim[1]: 199 vedo.logger.warning(f"Null range for Figure {self.title}... returning an empty Assembly.") 200 Assembly.__init__(self) 201 self.yscale = 0 202 return 203 204 if aspect == "equal": 205 self.aspect = dx / dy # so that yscale becomes 1 206 207 self.yscale = dx / dy / self.aspect 208 209 y0lim *= self.yscale 210 y1lim *= self.yscale 211 212 self.x0lim = x0lim 213 self.x1lim = x1lim 214 self.y0lim = y0lim 215 self.y1lim = y1lim 216 217 self.ztolerance = options.pop("ztolerance", None) 218 if self.ztolerance is None: 219 self.ztolerance = dx / 5000 220 221 ############## create axes 222 if self.axopts: 223 axes_opts = self.axopts 224 if self.axopts is True or self.axopts == 1: 225 axes_opts = {} 226 227 tp, ts = utils.make_ticks(y0lim / self.yscale, y1lim / self.yscale, number_of_divisions) 228 labs = [] 229 for i in range(1, len(tp) - 1): 230 ynew = utils.lin_interpolate(tp[i], [0, 1], [y0lim, y1lim]) 231 labs.append([ynew, ts[i]]) 232 233 if self.title: 234 axes_opts["htitle"] = self.title 235 axes_opts["y_values_and_labels"] = labs 236 axes_opts["xrange"] = (x0lim, x1lim) 237 axes_opts["yrange"] = (y0lim, y1lim) 238 axes_opts["zrange"] = (0, 0) 239 axes_opts["y_use_bounds"] = True 240 241 if "c" not in axes_opts and "ac" in options: 242 axes_opts["c"] = options["ac"] 243 244 self.axes = addons.Axes(**axes_opts) 245 246 Assembly.__init__(self, [self.axes]) 247 self.name = "Figure" 248 249 vedo.last_figure = self if settings.remember_last_figure_format else None 250 return
Create an empty formatted figure for plotting.
Arguments:
- xlim : (list) range of the x-axis as [x0, x1]
- ylim : (list) range of the y-axis as [y0, y1]
- aspect : (float, str)
the desired aspect ratio of the histogram. Default is 4/3.
Use
aspect="equal"to force the same units in x and y. - padding : (float, list) keep a padding space from the axes (as a fraction of the axis size). This can be a list of four numbers.
- xtitle : (str)
title for the x-axis, can also be set using
axes=dict(xtitle="my x axis") - ytitle : (str)
title for the y-axis, can also be set using
axes=dict(ytitle="my y axis") - grid : (bool)
show the background grid for the axes, can also be set using
axes=dict(xygrid=True) - axes : (dict)
an extra dictionary of options for the
vedo.addons.Axesobject
366 def insert(self, *objs, rescale=True, as3d=True, adjusted=False, cut=True): 367 """ 368 Insert objects into a Figure. 369 370 The recommended syntax is to use "+=", which calls `insert()` under the hood. 371 If a whole Figure is added with "+=", it is unpacked and its objects are added 372 one by one. 373 374 Arguments: 375 rescale : (bool) 376 rescale the y axis position while inserting the object. 377 as3d : (bool) 378 if True keep the aspect ratio of the 3d object, otherwise stretch it in y. 379 adjusted : (bool) 380 adjust the scaling according to the shortest axis 381 cut : (bool) 382 cut off the parts of the object which go beyond the axes frame. 383 """ 384 for a in objs: 385 386 if a in self.actors: 387 # should not add twice the same object in plot 388 continue 389 390 if isinstance(a, vedo.Points): # hacky way to identify Points 391 if a.ncells == a.npoints: 392 poly = a.polydata(False) 393 if poly.GetNumberOfPolys() == 0 and poly.GetNumberOfLines() == 0: 394 as3d = False 395 rescale = True 396 397 if isinstance(a, (shapes.Arrow, shapes.Arrow2D)): 398 # discard input Arrow and substitute it with a brand new one 399 # (because scaling would fatally distort the shape) 400 prop = a.GetProperty() 401 prop.LightingOff() 402 py = a.base[1] 403 a.top[1] = (a.top[1] - py) * self.yscale + py 404 b = shapes.Arrow2D(a.base, a.top, s=a.s, fill=a.fill).z(a.z()) 405 b.SetProperty(prop) 406 b.y(py * self.yscale) 407 a = b 408 409 # elif isinstance(a, shapes.Rectangle) and a.radius is not None: 410 # # discard input Rectangle and substitute it with a brand new one 411 # # (because scaling would fatally distort the shape of the corners) 412 # py = a.corner1[1] 413 # rx1,ry1,rz1 = a.corner1 414 # rx2,ry2,rz2 = a.corner2 415 # ry2 = (ry2-py) * self.yscale + py 416 # b = shapes.Rectangle([rx1,0,rz1], [rx2,ry2,rz2], radius=a.radius).z(a.z()) 417 # b.SetProperty(a.GetProperty()) 418 # b.y(py / self.yscale) 419 # a = b 420 421 else: 422 423 if rescale: 424 425 if not isinstance(a, Figure): 426 427 if as3d and not isinstance(a, self.force_scaling_types): 428 if adjusted: 429 scl = np.min([1, self.yscale]) 430 else: 431 scl = self.yscale 432 433 a.scale(scl) 434 435 else: 436 a.scale([1, self.yscale, 1]) 437 438 # shift it in y 439 a.y(a.y() * self.yscale) 440 441 if cut: 442 try: 443 bx0, bx1, by0, by1, _, _ = a.bounds() 444 if self.y0lim > by0: 445 a.cut_with_plane([0, self.y0lim, 0], [0, 1, 0]) 446 if self.y1lim < by1: 447 a.cut_with_plane([0, self.y1lim, 0], [0, -1, 0]) 448 if self.x0lim > bx0: 449 a.cut_with_plane([self.x0lim, 0, 0], [1, 0, 0]) 450 if self.x1lim < bx1: 451 a.cut_with_plane([self.x1lim, 0, 0], [-1, 0, 0]) 452 except: 453 # print("insert(): cannot cut", [a]) 454 pass 455 456 self.AddPart(a) 457 self.actors.append(a) 458 459 return self
Insert objects into a Figure.
The recommended syntax is to use "+=", which calls insert() under the hood.
If a whole Figure is added with "+=", it is unpacked and its objects are added
one by one.
Arguments:
- rescale : (bool) rescale the y axis position while inserting the object.
- as3d : (bool) if True keep the aspect ratio of the 3d object, otherwise stretch it in y.
- adjusted : (bool) adjust the scaling according to the shortest axis
- cut : (bool) cut off the parts of the object which go beyond the axes frame.
461 def add_label(self, text, c=None, marker="", mc="black"): 462 """ 463 Manually add en entry label to the legend. 464 465 Arguments: 466 text : (str) 467 text string for the label. 468 c : (str) 469 color of the text 470 marker : (str), Mesh 471 a marker char or a Mesh object to be used as marker 472 mc : (str) 473 color for the marker 474 """ 475 newlabel = LabelData() 476 newlabel.text = text.replace("\n", " ") 477 newlabel.tcolor = c 478 newlabel.marker = marker 479 newlabel.mcolor = mc 480 self.labels.append(newlabel) 481 return self
Manually add en entry label to the legend.
Arguments:
- text : (str) text string for the label.
- c : (str) color of the text
- marker : (str), Mesh a marker char or a Mesh object to be used as marker
- mc : (str) color for the marker
483 def add_legend( 484 self, 485 pos="top-right", 486 relative=True, 487 font=None, 488 s=1, 489 c=None, 490 vspace=1.75, 491 padding=0.1, 492 radius=0, 493 alpha=1, 494 bc="k7", 495 lw=1, 496 lc="k4", 497 z=0, 498 ): 499 """ 500 Add existing labels to form a legend box. 501 Labels have been previously filled with eg: `plot(..., label="text")` 502 503 Arguments: 504 pos : (str, list) 505 A string or 2D coordinates. The default is "top-right". 506 relative : (bool) 507 control whether `pos` is absolute or relative, e.i. normalized 508 to the x and y ranges so that x and y in `pos=[x,y]` should be 509 both in the range [0,1]. 510 This flag is ignored if a string despcriptor is passed. 511 Default is True. 512 font : (str, int) 513 font name or number. 514 Check [available fonts here](https://vedo.embl.es/fonts). 515 s : (float) 516 global size of the legend 517 c : (str) 518 color of the text 519 vspace : (float) 520 vertical spacing of lines 521 padding : (float) 522 padding of the box as a fraction of the text size 523 radius : (float) 524 border radius of the box 525 alpha : (float) 526 opacity of the box. Values below 1 may cause poor rendering 527 because of antialiasing. 528 Use alpha = 0 to remove the box. 529 bc : (str) 530 box color 531 lw : (int) 532 border line width of the box in pixel units 533 lc : (int) 534 border line color of the box 535 z : (float) 536 set the zorder as z position (useful to avoid overlap) 537 """ 538 sx = self.x1lim - self.x0lim 539 s = s * sx / 55 # so that input can be about 1 540 541 ds = 0 542 texts = [] 543 mks = [] 544 for i, t in enumerate(self.labels): 545 label = self.labels[i] 546 t = label.text 547 548 if label.tcolor is not None: 549 c = label.tcolor 550 551 tx = vedo.shapes.Text3D(t, s=s, c=c, justify="center-left", font=font) 552 y0, y1 = tx.ybounds() 553 ds = max(y1 - y0, ds) 554 texts.append(tx) 555 556 mk = label.marker 557 if isinstance(mk, vedo.Points): 558 mk = mk.clone(deep=False).lighting("off") 559 cm = mk.center_of_mass() 560 ty0, ty1 = tx.ybounds() 561 oby0, oby1 = mk.ybounds() 562 mk.shift(-cm) 563 mk.SetOrigin(cm) 564 mk.scale((ty1 - ty0) / (oby1 - oby0)) 565 mk.scale([1.1, 1.1, 0.01]) 566 elif mk == "-": 567 mk = vedo.shapes.Marker(mk, s=s * 2) 568 mk.color(label.mcolor) 569 else: 570 mk = vedo.shapes.Marker(mk, s=s) 571 mk.color(label.mcolor) 572 mks.append(mk) 573 574 for i, tx in enumerate(texts): 575 tx.shift(0, -(i + 0) * ds * vspace) 576 577 for i, mk in enumerate(mks): 578 mk.shift(-ds * 1.75, -(i + 0) * ds * vspace, 0) 579 580 acts = texts + mks 581 582 aleg = Assembly(acts) # .show(axes=1).close() 583 x0, x1, y0, y1, _, _ = aleg.GetBounds() 584 585 if alpha: 586 dx = x1 - x0 587 dy = y1 - y0 588 589 if not utils.is_sequence(padding): 590 padding = [padding] * 4 591 padding = min(padding) 592 padding = min(padding * dx, padding * dy) 593 if len(self.labels) == 1: 594 padding *= 4 595 x0 -= padding 596 x1 += padding 597 y0 -= padding 598 y1 += padding 599 600 box = shapes.Rectangle([x0, y0], [x1, y1], radius=radius, c=bc, alpha=alpha) 601 box.shift(0, 0, -dy / 100).pickable(False) 602 if lc: 603 box.lc(lc).lw(lw) 604 aleg.AddPart(box) 605 606 xlim = self.xlim 607 ylim = self.ylim 608 if isinstance(pos, str): 609 px, py = 0, 0 610 rx, ry = (xlim[1] + xlim[0]) / 2, (ylim[1] + ylim[0]) / 2 611 shx, shy = 0, 0 612 if "top" in pos: 613 if "cent" in pos: 614 px, py = rx, ylim[1] 615 shx, shy = (x0 + x1) / 2, y1 616 elif "left" in pos: 617 px, py = xlim[0], ylim[1] 618 shx, shy = x0, y1 619 else: # "right" 620 px, py = xlim[1], ylim[1] 621 shx, shy = x1, y1 622 elif "bot" in pos: 623 if "left" in pos: 624 px, py = xlim[0], ylim[0] 625 shx, shy = x0, y0 626 elif "right" in pos: 627 px, py = xlim[1], ylim[0] 628 shx, shy = x1, y0 629 else: # "cent" 630 px, py = rx, ylim[0] 631 shx, shy = (x0 + x1) / 2, y0 632 elif "cent" in pos: 633 if "left" in pos: 634 px, py = xlim[0], ry 635 shx, shy = x0, (y0 + y1) / 2 636 elif "right" in pos: 637 px, py = xlim[1], ry 638 shx, shy = x1, (y0 + y1) / 2 639 else: 640 vedo.logger.error(f"in add_legend(), cannot understand {pos}") 641 raise RuntimeError 642 643 else: 644 645 if relative: 646 rx, ry = pos[0], pos[1] 647 px = (xlim[1] - xlim[0]) * rx + xlim[0] 648 py = (ylim[1] - ylim[0]) * ry + ylim[0] 649 z *= xlim[1] - xlim[0] 650 else: 651 px, py = pos[0], pos[1] 652 shx, shy = x0, y1 653 654 aleg.pos(px - shx, py * self.yscale - shy, self.z() + sx / 50 + z) 655 656 self.insert(aleg, rescale=False, cut=False) 657 self.legend = aleg 658 aleg.name = "Legend" 659 return self
Add existing labels to form a legend box.
Labels have been previously filled with eg: plot(..., label="text")
Arguments:
- pos : (str, list) A string or 2D coordinates. The default is "top-right".
- relative : (bool)
control whether
posis absolute or relative, e.i. normalized to the x and y ranges so that x and y inpos=[x,y]should be both in the range [0,1]. This flag is ignored if a string despcriptor is passed. Default is True. - font : (str, int) font name or number. Check available fonts here.
- s : (float) global size of the legend
- c : (str) color of the text
- vspace : (float) vertical spacing of lines
- padding : (float) padding of the box as a fraction of the text size
- radius : (float) border radius of the box
- alpha : (float) opacity of the box. Values below 1 may cause poor rendering because of antialiasing. Use alpha = 0 to remove the box.
- bc : (str) box color
- lw : (int) border line width of the box in pixel units
- lc : (int) border line color of the box
- z : (float) set the zorder as z position (useful to avoid overlap)
661 def as2d(self, pos="bottom-left", scale=1, padding=0.05): 662 """ 663 Convert the Figure into a 2D static object (a 2D Assembly). 664 665 Arguments: 666 pos : (str, list) 667 position in 2D, as atring or list (x,y). 668 Any combination of "center", "top", "bottom", "left" and "right" will work. 669 The center of the renderer is [0,0] while top-right is [1,1]. 670 scale : (float) 671 scaling factor 672 padding : (float, list) 673 a single value or a list (xpad, ypad) 674 675 Returns: 676 `Group` object. 677 """ 678 x0, x1 = self.xbounds() 679 y0, y1 = self.ybounds() 680 pp = self.pos() 681 x0 -= pp[0] 682 x1 -= pp[0] 683 y0 -= pp[1] 684 y1 -= pp[1] 685 686 if not utils.is_sequence(padding): 687 padding = (padding, padding) 688 padding = np.array(padding) 689 690 if "cent" in pos: 691 offset = [(x0 + x1) / 2, (y0 + y1) / 2] 692 position = [0, 0] 693 if "right" in pos: 694 offset[0] = x1 695 position = [1 - padding[0], 0] 696 if "left" in pos: 697 offset[0] = x0 698 position = [-1 + padding[0], 0] 699 if "top" in pos: 700 offset[1] = y1 701 position = [0, 1 - padding[1]] 702 if "bottom" in pos: 703 offset[1] = y0 704 position = [0, -1 + padding[1]] 705 elif "top" in pos: 706 if "right" in pos: 707 offset = [x1, y1] 708 position = [1, 1] - padding 709 elif "left" in pos: 710 offset = [x0, y1] 711 position = [-1 + padding[0], 1 - padding[1]] 712 else: 713 raise ValueError(f"incomplete position pos='{pos}'") 714 elif "bottom" in pos: 715 if "right" in pos: 716 offset = [x1, y0] 717 position = [1 - padding[0], -1 + padding[1]] 718 elif "left" in pos: 719 offset = [x0, y0] 720 position = [-1, -1] + padding 721 else: 722 raise ValueError(f"incomplete position pos='{pos}'") 723 else: 724 offset = [0, 0] 725 position = pos 726 727 scanned = [] 728 group = Group() 729 for a in self.recursive_unpack(): 730 if a in scanned: 731 continue 732 if not isinstance(a, vedo.Points): 733 continue 734 if a.npoints == 0: 735 continue 736 if a.GetProperty().GetRepresentation() == 1: 737 # wireframe is not rendered correctly in 2d 738 continue 739 a2d = _to2d(a, offset, scale * 550 / (x1 - x0)) 740 a2d.SetPosition(position) 741 group += a2d 742 return group
Convert the Figure into a 2D static object (a 2D Assembly).
Arguments:
- pos : (str, list) position in 2D, as atring or list (x,y). Any combination of "center", "top", "bottom", "left" and "right" will work. The center of the renderer is [0,0] while top-right is [1,1].
- scale : (float) scaling factor
- padding : (float, list) a single value or a list (xpad, ypad)
Returns:
Groupobject.
Inherited Members
746class Histogram1D(Figure): 747 "1D histogramming." 748 749 def __init__( 750 self, 751 data, 752 weights=None, 753 bins=None, 754 errors=False, 755 density=False, 756 logscale=False, 757 fill=True, 758 radius=0.075, 759 c="olivedrab", 760 gap=0.02, 761 alpha=1, 762 outline=False, 763 lw=2, 764 lc="k", 765 texture="", 766 marker="", 767 ms=None, 768 mc=None, 769 ma=None, 770 # Figure and axes options: 771 like=None, 772 xlim=None, 773 ylim=(0, None), 774 aspect=4 / 3, 775 padding=(0.0, 0.0, 0.0, 0.05), 776 title="", 777 xtitle=" ", 778 ytitle=" ", 779 ac="k", 780 grid=False, 781 ztolerance=None, 782 label="", 783 **fig_kwargs, 784 ): 785 """ 786 Creates a `Histogram1D(Figure)` object. 787 788 Arguments: 789 weights : (list) 790 An array of weights, of the same shape as `data`. Each value in `data` 791 only contributes its associated weight towards the bin count (instead of 1). 792 bins : (int) 793 number of bins 794 density : (bool) 795 normalize the area to 1 by dividing by the nr of entries and bin size 796 logscale : (bool) 797 use logscale on y-axis 798 fill : (bool) 799 fill bars with solid color `c` 800 gap : (float) 801 leave a small space btw bars 802 radius : (float) 803 border radius of the top of the histogram bar. Default value is 0.1. 804 texture : (str) 805 url or path to an image to be used as texture for the bin 806 outline : (bool) 807 show outline of the bins 808 errors : (bool) 809 show error bars 810 xtitle : (str) 811 title for the x-axis, can also be set using `axes=dict(xtitle="my x axis")` 812 ytitle : (str) 813 title for the y-axis, can also be set using `axes=dict(ytitle="my y axis")` 814 padding : (float), list 815 keep a padding space from the axes (as a fraction of the axis size). 816 This can be a list of four numbers. 817 aspect : (float) 818 the desired aspect ratio of the histogram. Default is 4/3. 819 grid : (bool) 820 show the background grid for the axes, can also be set using `axes=dict(xygrid=True)` 821 ztolerance : (float) 822 a tolerance factor to superimpose objects (along the z-axis). 823 824 Examples: 825 - [histo_1d_a.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_1d_a.py) 826 - [histo_1d_b.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_1d_b.py) 827 - [histo_1d_c.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_1d_c.py) 828 - [histo_1d_d.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_1d_d.py) 829 830  831 """ 832 833 # purge NaN from data 834 valid_ids = np.all(np.logical_not(np.isnan(data))) 835 data = np.asarray(data[valid_ids]).ravel() 836 837 # if data.dtype is integer try to center bins by default 838 if like is None and bins is None and np.issubdtype(data.dtype, np.integer): 839 if xlim is None and ylim == (0, None): 840 x1, x0 = data.max(), data.min() 841 if 0 < x1 - x0 <= 100: 842 bins = x1 - x0 + 1 843 xlim = (x0 - 0.5, x1 + 0.5) 844 845 if like is None and vedo.last_figure is not None: 846 if xlim is None and ylim == (0, None): 847 like = vedo.last_figure 848 849 if like is not None: 850 xlim = like.xlim 851 ylim = like.ylim 852 aspect = like.aspect 853 padding = like.padding 854 if bins is None: 855 bins = like.bins 856 if bins is None: 857 bins = 20 858 859 if utils.is_sequence(xlim): 860 # deal with user passing eg [x0, None] 861 _x0, _x1 = xlim 862 if _x0 is None: 863 _x0 = data.min() 864 if _x1 is None: 865 _x1 = data.max() 866 xlim = [_x0, _x1] 867 868 fs, edges = np.histogram(data, bins=bins, weights=weights, range=xlim) 869 binsize = edges[1] - edges[0] 870 ntot = data.shape[0] 871 872 fig_kwargs["title"] = title 873 fig_kwargs["xtitle"] = xtitle 874 fig_kwargs["ytitle"] = ytitle 875 fig_kwargs["ac"] = ac 876 fig_kwargs["ztolerance"] = ztolerance 877 fig_kwargs["grid"] = grid 878 879 unscaled_errors = np.sqrt(fs) 880 if density: 881 scaled_errors = unscaled_errors / (ntot * binsize) 882 fs = fs / (ntot * binsize) 883 if ytitle == " ": 884 ytitle = f"counts / ({ntot} x {utils.precision(binsize,3)})" 885 fig_kwargs["ytitle"] = ytitle 886 elif logscale: 887 se_up = np.log10(fs + unscaled_errors / 2 + 1) 888 se_dw = np.log10(fs - unscaled_errors / 2 + 1) 889 scaled_errors = np.c_[se_up, se_dw] 890 fs = np.log10(fs + 1) 891 if ytitle == " ": 892 ytitle = "log_10 (counts+1)" 893 fig_kwargs["ytitle"] = ytitle 894 895 x0, x1 = np.min(edges), np.max(edges) 896 y0, y1 = ylim[0], np.max(fs) 897 898 _errors = [] 899 if errors: 900 if density: 901 y1 += max(scaled_errors) / 2 902 _errors = scaled_errors 903 elif logscale: 904 y1 = max(scaled_errors[:, 0]) 905 _errors = scaled_errors 906 else: 907 y1 += max(unscaled_errors) / 2 908 _errors = unscaled_errors 909 910 if like is None: 911 ylim = list(ylim) 912 if xlim is None: 913 xlim = [x0, x1] 914 if ylim[1] is None: 915 ylim[1] = y1 916 if ylim[0] != 0: 917 ylim[0] = y0 918 919 self.title = title 920 self.xtitle = xtitle 921 self.ytitle = ytitle 922 self.entries = ntot 923 self.frequencies = fs 924 self.errors = _errors 925 self.edges = edges 926 self.centers = (edges[0:-1] + edges[1:]) / 2 927 self.mean = data.mean() 928 self.std = data.std() 929 self.bins = edges # internally used by "like" 930 931 ############################### stats legend as htitle 932 addstats = False 933 if not title: 934 if "axes" not in fig_kwargs: 935 addstats = True 936 axes_opts = {} 937 fig_kwargs["axes"] = axes_opts 938 elif fig_kwargs["axes"] is False: 939 pass 940 else: 941 axes_opts = fig_kwargs["axes"] 942 if "htitle" not in axes_opts: 943 addstats = True 944 945 if addstats: 946 htitle = f"Entries:~~{int(self.entries)} " 947 htitle += f"Mean:~~{utils.precision(self.mean, 4)} " 948 htitle += f"STD:~~{utils.precision(self.std, 4)} " 949 950 axes_opts["htitle"] = htitle 951 axes_opts["htitle_justify"] = "bottom-left" 952 axes_opts["htitle_size"] = 0.016 953 axes_opts["htitle_offset"] = [-0.49, 0.01, 0] 954 955 if mc is None: 956 mc = lc 957 if ma is None: 958 ma = alpha 959 960 if label: 961 nlab = LabelData() 962 nlab.text = label 963 nlab.tcolor = ac 964 nlab.marker = marker 965 nlab.mcolor = mc 966 if not marker: 967 nlab.marker = "s" 968 nlab.mcolor = c 969 fig_kwargs["label"] = nlab 970 971 ############################################### Figure init 972 Figure.__init__(self, xlim, ylim, aspect, padding, **fig_kwargs) 973 if not self.yscale: 974 return 975 976 if utils.is_sequence(bins): 977 myedges = np.array(bins) 978 bins = len(bins) - 1 979 else: 980 myedges = edges 981 982 bin_centers = [] 983 for i in range(bins): 984 x = (myedges[i] + myedges[i + 1]) / 2 985 bin_centers.append([x, fs[i], 0]) 986 987 rs = [] 988 maxheigth = 0 989 if not fill and not outline and not errors and not marker: 990 outline = True # otherwise it's empty.. 991 992 if fill: ##################### 993 if outline: 994 gap = 0 995 996 for i in range(bins): 997 F = fs[i] 998 if not F: 999 continue 1000 p0 = (myedges[i] + gap * binsize, 0, 0) 1001 p1 = (myedges[i + 1] - gap * binsize, F, 0) 1002 1003 if radius: 1004 if gap: 1005 rds = np.array([0, 0, radius, radius]) 1006 else: 1007 rd1 = 0 if i < bins - 1 and fs[i + 1] >= F else radius / 2 1008 rd2 = 0 if i > 0 and fs[i - 1] >= F else radius / 2 1009 rds = np.array([0, 0, rd1, rd2]) 1010 p1_yscaled = [p1[0], p1[1] * self.yscale, 0] 1011 r = shapes.Rectangle(p0, p1_yscaled, radius=rds * binsize, res=6) 1012 r.scale([1, 1 / self.yscale, 1]) 1013 r.radius = None # so it doesnt get recreated and rescaled by insert() 1014 else: 1015 r = shapes.Rectangle(p0, p1) 1016 1017 if texture: 1018 r.texture(texture) 1019 c = "w" 1020 # if texture: # causes Segmentation fault vtk9.0.3 1021 # if i>0 and rs[i-1].GetTexture(): # reuse the same texture obj 1022 # r.texture(rs[i-1].GetTexture()) 1023 # else: 1024 # r.texture(texture) 1025 # c = 'w' 1026 1027 r.PickableOff() 1028 maxheigth = max(maxheigth, p1[1]) 1029 if c in colors.cmaps_names: 1030 col = colors.color_map((p0[0] + p1[0]) / 2, c, myedges[0], myedges[-1]) 1031 else: 1032 col = c 1033 r.color(col).alpha(alpha).lighting("off") 1034 r.z(self.ztolerance) 1035 rs.append(r) 1036 1037 if outline: ##################### 1038 lns = [[myedges[0], 0, 0]] 1039 for i in range(bins): 1040 lns.append([myedges[i], fs[i], 0]) 1041 lns.append([myedges[i + 1], fs[i], 0]) 1042 maxheigth = max(maxheigth, fs[i]) 1043 lns.append([myedges[-1], 0, 0]) 1044 outl = shapes.Line(lns, c=lc, alpha=alpha, lw=lw) 1045 outl.z(self.ztolerance * 2) 1046 rs.append(outl) 1047 1048 if errors: ##################### 1049 for i in range(bins): 1050 x = self.centers[i] 1051 f = fs[i] 1052 if not f: 1053 continue 1054 err = _errors[i] 1055 if utils.is_sequence(err): 1056 el = shapes.Line([x, err[0], 0], [x, err[1], 0], c=lc, alpha=alpha, lw=lw) 1057 else: 1058 el = shapes.Line( 1059 [x, f - err / 2, 0], [x, f + err / 2, 0], c=lc, alpha=alpha, lw=lw 1060 ) 1061 el.z(self.ztolerance * 3) 1062 rs.append(el) 1063 1064 if marker: ##################### 1065 1066 # remove empty bins (we dont want a marker there) 1067 bin_centers = np.array(bin_centers) 1068 bin_centers = bin_centers[bin_centers[:, 1] > 0] 1069 1070 if utils.is_sequence(ms): ### variable point size 1071 mk = shapes.Marker(marker, s=1) 1072 mk.scale([1, 1 / self.yscale, 1]) 1073 msv = np.zeros_like(bin_centers) 1074 msv[:, 0] = ms 1075 marked = shapes.Glyph( 1076 bin_centers, mk, c=mc, orientation_array=msv, scale_by_vector_size=True 1077 ) 1078 else: ### fixed point size 1079 1080 if ms is None: 1081 ms = (xlim[1] - xlim[0]) / 100.0 1082 else: 1083 ms = (xlim[1] - xlim[0]) / 100.0 * ms 1084 1085 if utils.is_sequence(mc): 1086 mk = shapes.Marker(marker, s=ms) 1087 mk.scale([1, 1 / self.yscale, 1]) 1088 msv = np.zeros_like(bin_centers) 1089 msv[:, 0] = 1 1090 marked = shapes.Glyph( 1091 bin_centers, mk, c=mc, orientation_array=msv, scale_by_vector_size=True 1092 ) 1093 else: 1094 mk = shapes.Marker(marker, s=ms) 1095 mk.scale([1, 1 / self.yscale, 1]) 1096 marked = shapes.Glyph(bin_centers, mk, c=mc) 1097 1098 marked.alpha(ma) 1099 marked.z(self.ztolerance * 4) 1100 rs.append(marked) 1101 1102 self.insert(*rs, as3d=False) 1103 self.name = "Histogram1D" 1104 1105 def print(self, **kwargs): 1106 """Print infos about this histogram""" 1107 txt = ( 1108 f"{self.name} {self.title}\n" 1109 f" xtitle = '{self.xtitle}'\n" 1110 f" ytitle = '{self.ytitle}'\n" 1111 f" entries = {self.entries}\n" 1112 f" mean = {self.mean}\n" 1113 f" std = {self.std}" 1114 ) 1115 colors.printc(txt, **kwargs)
1D histogramming.
749 def __init__( 750 self, 751 data, 752 weights=None, 753 bins=None, 754 errors=False, 755 density=False, 756 logscale=False, 757 fill=True, 758 radius=0.075, 759 c="olivedrab", 760 gap=0.02, 761 alpha=1, 762 outline=False, 763 lw=2, 764 lc="k", 765 texture="", 766 marker="", 767 ms=None, 768 mc=None, 769 ma=None, 770 # Figure and axes options: 771 like=None, 772 xlim=None, 773 ylim=(0, None), 774 aspect=4 / 3, 775 padding=(0.0, 0.0, 0.0, 0.05), 776 title="", 777 xtitle=" ", 778 ytitle=" ", 779 ac="k", 780 grid=False, 781 ztolerance=None, 782 label="", 783 **fig_kwargs, 784 ): 785 """ 786 Creates a `Histogram1D(Figure)` object. 787 788 Arguments: 789 weights : (list) 790 An array of weights, of the same shape as `data`. Each value in `data` 791 only contributes its associated weight towards the bin count (instead of 1). 792 bins : (int) 793 number of bins 794 density : (bool) 795 normalize the area to 1 by dividing by the nr of entries and bin size 796 logscale : (bool) 797 use logscale on y-axis 798 fill : (bool) 799 fill bars with solid color `c` 800 gap : (float) 801 leave a small space btw bars 802 radius : (float) 803 border radius of the top of the histogram bar. Default value is 0.1. 804 texture : (str) 805 url or path to an image to be used as texture for the bin 806 outline : (bool) 807 show outline of the bins 808 errors : (bool) 809 show error bars 810 xtitle : (str) 811 title for the x-axis, can also be set using `axes=dict(xtitle="my x axis")` 812 ytitle : (str) 813 title for the y-axis, can also be set using `axes=dict(ytitle="my y axis")` 814 padding : (float), list 815 keep a padding space from the axes (as a fraction of the axis size). 816 This can be a list of four numbers. 817 aspect : (float) 818 the desired aspect ratio of the histogram. Default is 4/3. 819 grid : (bool) 820 show the background grid for the axes, can also be set using `axes=dict(xygrid=True)` 821 ztolerance : (float) 822 a tolerance factor to superimpose objects (along the z-axis). 823 824 Examples: 825 - [histo_1d_a.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_1d_a.py) 826 - [histo_1d_b.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_1d_b.py) 827 - [histo_1d_c.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_1d_c.py) 828 - [histo_1d_d.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_1d_d.py) 829 830  831 """ 832 833 # purge NaN from data 834 valid_ids = np.all(np.logical_not(np.isnan(data))) 835 data = np.asarray(data[valid_ids]).ravel() 836 837 # if data.dtype is integer try to center bins by default 838 if like is None and bins is None and np.issubdtype(data.dtype, np.integer): 839 if xlim is None and ylim == (0, None): 840 x1, x0 = data.max(), data.min() 841 if 0 < x1 - x0 <= 100: 842 bins = x1 - x0 + 1 843 xlim = (x0 - 0.5, x1 + 0.5) 844 845 if like is None and vedo.last_figure is not None: 846 if xlim is None and ylim == (0, None): 847 like = vedo.last_figure 848 849 if like is not None: 850 xlim = like.xlim 851 ylim = like.ylim 852 aspect = like.aspect 853 padding = like.padding 854 if bins is None: 855 bins = like.bins 856 if bins is None: 857 bins = 20 858 859 if utils.is_sequence(xlim): 860 # deal with user passing eg [x0, None] 861 _x0, _x1 = xlim 862 if _x0 is None: 863 _x0 = data.min() 864 if _x1 is None: 865 _x1 = data.max() 866 xlim = [_x0, _x1] 867 868 fs, edges = np.histogram(data, bins=bins, weights=weights, range=xlim) 869 binsize = edges[1] - edges[0] 870 ntot = data.shape[0] 871 872 fig_kwargs["title"] = title 873 fig_kwargs["xtitle"] = xtitle 874 fig_kwargs["ytitle"] = ytitle 875 fig_kwargs["ac"] = ac 876 fig_kwargs["ztolerance"] = ztolerance 877 fig_kwargs["grid"] = grid 878 879 unscaled_errors = np.sqrt(fs) 880 if density: 881 scaled_errors = unscaled_errors / (ntot * binsize) 882 fs = fs / (ntot * binsize) 883 if ytitle == " ": 884 ytitle = f"counts / ({ntot} x {utils.precision(binsize,3)})" 885 fig_kwargs["ytitle"] = ytitle 886 elif logscale: 887 se_up = np.log10(fs + unscaled_errors / 2 + 1) 888 se_dw = np.log10(fs - unscaled_errors / 2 + 1) 889 scaled_errors = np.c_[se_up, se_dw] 890 fs = np.log10(fs + 1) 891 if ytitle == " ": 892 ytitle = "log_10 (counts+1)" 893 fig_kwargs["ytitle"] = ytitle 894 895 x0, x1 = np.min(edges), np.max(edges) 896 y0, y1 = ylim[0], np.max(fs) 897 898 _errors = [] 899 if errors: 900 if density: 901 y1 += max(scaled_errors) / 2 902 _errors = scaled_errors 903 elif logscale: 904 y1 = max(scaled_errors[:, 0]) 905 _errors = scaled_errors 906 else: 907 y1 += max(unscaled_errors) / 2 908 _errors = unscaled_errors 909 910 if like is None: 911 ylim = list(ylim) 912 if xlim is None: 913 xlim = [x0, x1] 914 if ylim[1] is None: 915 ylim[1] = y1 916 if ylim[0] != 0: 917 ylim[0] = y0 918 919 self.title = title 920 self.xtitle = xtitle 921 self.ytitle = ytitle 922 self.entries = ntot 923 self.frequencies = fs 924 self.errors = _errors 925 self.edges = edges 926 self.centers = (edges[0:-1] + edges[1:]) / 2 927 self.mean = data.mean() 928 self.std = data.std() 929 self.bins = edges # internally used by "like" 930 931 ############################### stats legend as htitle 932 addstats = False 933 if not title: 934 if "axes" not in fig_kwargs: 935 addstats = True 936 axes_opts = {} 937 fig_kwargs["axes"] = axes_opts 938 elif fig_kwargs["axes"] is False: 939 pass 940 else: 941 axes_opts = fig_kwargs["axes"] 942 if "htitle" not in axes_opts: 943 addstats = True 944 945 if addstats: 946 htitle = f"Entries:~~{int(self.entries)} " 947 htitle += f"Mean:~~{utils.precision(self.mean, 4)} " 948 htitle += f"STD:~~{utils.precision(self.std, 4)} " 949 950 axes_opts["htitle"] = htitle 951 axes_opts["htitle_justify"] = "bottom-left" 952 axes_opts["htitle_size"] = 0.016 953 axes_opts["htitle_offset"] = [-0.49, 0.01, 0] 954 955 if mc is None: 956 mc = lc 957 if ma is None: 958 ma = alpha 959 960 if label: 961 nlab = LabelData() 962 nlab.text = label 963 nlab.tcolor = ac 964 nlab.marker = marker 965 nlab.mcolor = mc 966 if not marker: 967 nlab.marker = "s" 968 nlab.mcolor = c 969 fig_kwargs["label"] = nlab 970 971 ############################################### Figure init 972 Figure.__init__(self, xlim, ylim, aspect, padding, **fig_kwargs) 973 if not self.yscale: 974 return 975 976 if utils.is_sequence(bins): 977 myedges = np.array(bins) 978 bins = len(bins) - 1 979 else: 980 myedges = edges 981 982 bin_centers = [] 983 for i in range(bins): 984 x = (myedges[i] + myedges[i + 1]) / 2 985 bin_centers.append([x, fs[i], 0]) 986 987 rs = [] 988 maxheigth = 0 989 if not fill and not outline and not errors and not marker: 990 outline = True # otherwise it's empty.. 991 992 if fill: ##################### 993 if outline: 994 gap = 0 995 996 for i in range(bins): 997 F = fs[i] 998 if not F: 999 continue 1000 p0 = (myedges[i] + gap * binsize, 0, 0) 1001 p1 = (myedges[i + 1] - gap * binsize, F, 0) 1002 1003 if radius: 1004 if gap: 1005 rds = np.array([0, 0, radius, radius]) 1006 else: 1007 rd1 = 0 if i < bins - 1 and fs[i + 1] >= F else radius / 2 1008 rd2 = 0 if i > 0 and fs[i - 1] >= F else radius / 2 1009 rds = np.array([0, 0, rd1, rd2]) 1010 p1_yscaled = [p1[0], p1[1] * self.yscale, 0] 1011 r = shapes.Rectangle(p0, p1_yscaled, radius=rds * binsize, res=6) 1012 r.scale([1, 1 / self.yscale, 1]) 1013 r.radius = None # so it doesnt get recreated and rescaled by insert() 1014 else: 1015 r = shapes.Rectangle(p0, p1) 1016 1017 if texture: 1018 r.texture(texture) 1019 c = "w" 1020 # if texture: # causes Segmentation fault vtk9.0.3 1021 # if i>0 and rs[i-1].GetTexture(): # reuse the same texture obj 1022 # r.texture(rs[i-1].GetTexture()) 1023 # else: 1024 # r.texture(texture) 1025 # c = 'w' 1026 1027 r.PickableOff() 1028 maxheigth = max(maxheigth, p1[1]) 1029 if c in colors.cmaps_names: 1030 col = colors.color_map((p0[0] + p1[0]) / 2, c, myedges[0], myedges[-1]) 1031 else: 1032 col = c 1033 r.color(col).alpha(alpha).lighting("off") 1034 r.z(self.ztolerance) 1035 rs.append(r) 1036 1037 if outline: ##################### 1038 lns = [[myedges[0], 0, 0]] 1039 for i in range(bins): 1040 lns.append([myedges[i], fs[i], 0]) 1041 lns.append([myedges[i + 1], fs[i], 0]) 1042 maxheigth = max(maxheigth, fs[i]) 1043 lns.append([myedges[-1], 0, 0]) 1044 outl = shapes.Line(lns, c=lc, alpha=alpha, lw=lw) 1045 outl.z(self.ztolerance * 2) 1046 rs.append(outl) 1047 1048 if errors: ##################### 1049 for i in range(bins): 1050 x = self.centers[i] 1051 f = fs[i] 1052 if not f: 1053 continue 1054 err = _errors[i] 1055 if utils.is_sequence(err): 1056 el = shapes.Line([x, err[0], 0], [x, err[1], 0], c=lc, alpha=alpha, lw=lw) 1057 else: 1058 el = shapes.Line( 1059 [x, f - err / 2, 0], [x, f + err / 2, 0], c=lc, alpha=alpha, lw=lw 1060 ) 1061 el.z(self.ztolerance * 3) 1062 rs.append(el) 1063 1064 if marker: ##################### 1065 1066 # remove empty bins (we dont want a marker there) 1067 bin_centers = np.array(bin_centers) 1068 bin_centers = bin_centers[bin_centers[:, 1] > 0] 1069 1070 if utils.is_sequence(ms): ### variable point size 1071 mk = shapes.Marker(marker, s=1) 1072 mk.scale([1, 1 / self.yscale, 1]) 1073 msv = np.zeros_like(bin_centers) 1074 msv[:, 0] = ms 1075 marked = shapes.Glyph( 1076 bin_centers, mk, c=mc, orientation_array=msv, scale_by_vector_size=True 1077 ) 1078 else: ### fixed point size 1079 1080 if ms is None: 1081 ms = (xlim[1] - xlim[0]) / 100.0 1082 else: 1083 ms = (xlim[1] - xlim[0]) / 100.0 * ms 1084 1085 if utils.is_sequence(mc): 1086 mk = shapes.Marker(marker, s=ms) 1087 mk.scale([1, 1 / self.yscale, 1]) 1088 msv = np.zeros_like(bin_centers) 1089 msv[:, 0] = 1 1090 marked = shapes.Glyph( 1091 bin_centers, mk, c=mc, orientation_array=msv, scale_by_vector_size=True 1092 ) 1093 else: 1094 mk = shapes.Marker(marker, s=ms) 1095 mk.scale([1, 1 / self.yscale, 1]) 1096 marked = shapes.Glyph(bin_centers, mk, c=mc) 1097 1098 marked.alpha(ma) 1099 marked.z(self.ztolerance * 4) 1100 rs.append(marked) 1101 1102 self.insert(*rs, as3d=False) 1103 self.name = "Histogram1D"
Creates a Histogram1D(Figure) object.
Arguments:
- weights : (list)
An array of weights, of the same shape as
data. Each value indataonly contributes its associated weight towards the bin count (instead of 1). - bins : (int) number of bins
- density : (bool) normalize the area to 1 by dividing by the nr of entries and bin size
- logscale : (bool) use logscale on y-axis
- fill : (bool)
fill bars with solid color
c - gap : (float) leave a small space btw bars
- radius : (float) border radius of the top of the histogram bar. Default value is 0.1.
- texture : (str) url or path to an image to be used as texture for the bin
- outline : (bool) show outline of the bins
- errors : (bool) show error bars
- xtitle : (str)
title for the x-axis, can also be set using
axes=dict(xtitle="my x axis") - ytitle : (str)
title for the y-axis, can also be set using
axes=dict(ytitle="my y axis") - padding : (float), list keep a padding space from the axes (as a fraction of the axis size). This can be a list of four numbers.
- aspect : (float) the desired aspect ratio of the histogram. Default is 4/3.
- grid : (bool)
show the background grid for the axes, can also be set using
axes=dict(xygrid=True) - ztolerance : (float) a tolerance factor to superimpose objects (along the z-axis).
Examples:
1105 def print(self, **kwargs): 1106 """Print infos about this histogram""" 1107 txt = ( 1108 f"{self.name} {self.title}\n" 1109 f" xtitle = '{self.xtitle}'\n" 1110 f" ytitle = '{self.ytitle}'\n" 1111 f" entries = {self.entries}\n" 1112 f" mean = {self.mean}\n" 1113 f" std = {self.std}" 1114 ) 1115 colors.printc(txt, **kwargs)
Print infos about this histogram
Inherited Members
1119class Histogram2D(Figure): 1120 """2D histogramming.""" 1121 1122 def __init__( 1123 self, 1124 xvalues, 1125 yvalues=None, 1126 bins=25, 1127 weights=None, 1128 cmap="cividis", 1129 alpha=1, 1130 gap=0, 1131 scalarbar=True, 1132 # Figure and axes options: 1133 like=None, 1134 xlim=None, 1135 ylim=(None, None), 1136 zlim=(None, None), 1137 aspect=1, 1138 title="", 1139 xtitle=" ", 1140 ytitle=" ", 1141 ztitle="", 1142 ac="k", 1143 **fig_kwargs, 1144 ): 1145 """ 1146 Input data formats `[(x1,x2,..), (y1,y2,..)] or [(x1,y1), (x2,y2),..]` 1147 are both valid. 1148 1149 Use keyword `like=...` if you want to use the same format of a previously 1150 created Figure (useful when superimposing Figures) to make sure 1151 they are compatible and comparable. If they are not compatible 1152 you will receive an error message. 1153 1154 Arguments: 1155 bins : (list) 1156 binning as (nx, ny) 1157 weights : (list) 1158 array of weights to assign to each entry 1159 cmap : (str, lookuptable) 1160 color map name or look up table 1161 alpha : (float) 1162 opacity of the histogram 1163 gap : (float) 1164 separation between adjacent bins as a fraction for their size 1165 scalarbar : (bool) 1166 add a scalarbar to right of the histogram 1167 like : (Figure) 1168 grab and use the same format of the given Figure (for superimposing) 1169 xlim : (list) 1170 [x0, x1] range of interest. If left to None will automatically 1171 choose the minimum or the maximum of the data range. 1172 Data outside the range are completely ignored. 1173 ylim : (list) 1174 [y0, y1] range of interest. If left to None will automatically 1175 choose the minimum or the maximum of the data range. 1176 Data outside the range are completely ignored. 1177 aspect : (float) 1178 the desired aspect ratio of the figure. 1179 title : (str) 1180 title of the plot to appear on top. 1181 If left blank some statistics will be shown. 1182 xtitle : (str) 1183 x axis title 1184 ytitle : (str) 1185 y axis title 1186 ztitle : (str) 1187 title for the scalar bar 1188 ac : (str) 1189 axes color, additional keyword for Axes can also be added 1190 using e.g. `axes=dict(xygrid=True)` 1191 1192 Examples: 1193 - [histo_2d_a.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_2d_a.py) 1194 - [histo_2d_b.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_2d_b.py) 1195 1196  1197 """ 1198 xvalues = np.asarray(xvalues) 1199 if yvalues is None: 1200 # assume [(x1,y1), (x2,y2) ...] format 1201 yvalues = xvalues[:, 1] 1202 xvalues = xvalues[:, 0] 1203 else: 1204 yvalues = np.asarray(yvalues) 1205 1206 padding = [0, 0, 0, 0] 1207 1208 if like is None and vedo.last_figure is not None: 1209 if xlim is None and ylim == (None, None) and zlim == (None, None): 1210 like = vedo.last_figure 1211 1212 if like is not None: 1213 xlim = like.xlim 1214 ylim = like.ylim 1215 aspect = like.aspect 1216 padding = like.padding 1217 if bins is None: 1218 bins = like.bins 1219 if bins is None: 1220 bins = 20 1221 1222 if isinstance(bins, int): 1223 bins = (bins, bins) 1224 1225 if utils.is_sequence(xlim): 1226 # deal with user passing eg [x0, None] 1227 _x0, _x1 = xlim 1228 if _x0 is None: 1229 _x0 = xvalues.min() 1230 if _x1 is None: 1231 _x1 = xvalues.max() 1232 xlim = [_x0, _x1] 1233 1234 if utils.is_sequence(ylim): 1235 # deal with user passing eg [x0, None] 1236 _y0, _y1 = ylim 1237 if _y0 is None: 1238 _y0 = yvalues.min() 1239 if _y1 is None: 1240 _y1 = yvalues.max() 1241 ylim = [_y0, _y1] 1242 1243 H, xedges, yedges = np.histogram2d( 1244 xvalues, yvalues, weights=weights, bins=bins, range=(xlim, ylim) 1245 ) 1246 1247 xlim = np.min(xedges), np.max(xedges) 1248 ylim = np.min(yedges), np.max(yedges) 1249 dx, dy = xlim[1] - xlim[0], ylim[1] - ylim[0] 1250 1251 fig_kwargs["title"] = title 1252 fig_kwargs["xtitle"] = xtitle 1253 fig_kwargs["ytitle"] = ytitle 1254 fig_kwargs["ac"] = ac 1255 1256 self.entries = len(xvalues) 1257 self.frequencies = H 1258 self.edges = (xedges, yedges) 1259 self.mean = (xvalues.mean(), yvalues.mean()) 1260 self.std = (xvalues.std(), yvalues.std()) 1261 self.bins = bins # internally used by "like" 1262 1263 ############################### stats legend as htitle 1264 addstats = False 1265 if not title: 1266 if "axes" not in fig_kwargs: 1267 addstats = True 1268 axes_opts = {} 1269 fig_kwargs["axes"] = axes_opts 1270 elif fig_kwargs["axes"] is False: 1271 pass 1272 else: 1273 axes_opts = fig_kwargs["axes"] 1274 if "htitle" not in fig_kwargs["axes"]: 1275 addstats = True 1276 1277 if addstats: 1278 htitle = f"Entries:~~{int(self.entries)} " 1279 htitle += f"Mean:~~{utils.precision(self.mean, 3)} " 1280 htitle += f"STD:~~{utils.precision(self.std, 3)} " 1281 axes_opts["htitle"] = htitle 1282 axes_opts["htitle_justify"] = "bottom-left" 1283 axes_opts["htitle_size"] = 0.0175 1284 axes_opts["htitle_offset"] = [-0.49, 0.01, 0] 1285 1286 ############################################### Figure init 1287 Figure.__init__(self, xlim, ylim, aspect, padding, **fig_kwargs) 1288 1289 if self.yscale: 1290 ##################### the grid 1291 acts = [] 1292 g = shapes.Grid( 1293 pos=[(xlim[0] + xlim[1]) / 2, (ylim[0] + ylim[1]) / 2, 0], s=(dx, dy), res=bins[:2] 1294 ) 1295 g.alpha(alpha).lw(0).wireframe(False).flat().lighting("off") 1296 g.cmap(cmap, np.ravel(H.T), on="cells", vmin=zlim[0], vmax=zlim[1]) 1297 if gap: 1298 g.shrink(abs(1 - gap)) 1299 1300 if scalarbar: 1301 sc = g.add_scalarbar3d(ztitle, c=ac).scalarbar 1302 sc.scale([self.yscale, 1, 1]) ## prescale trick 1303 sbnds = sc.xbounds() 1304 sc.x(self.x1lim + (sbnds[1] - sbnds[0]) * 0.75) 1305 acts.append(sc) 1306 acts.append(g) 1307 1308 self.insert(*acts, as3d=False) 1309 self.name = "Histogram2D"
2D histogramming.
1122 def __init__( 1123 self, 1124 xvalues, 1125 yvalues=None, 1126 bins=25, 1127 weights=None, 1128 cmap="cividis", 1129 alpha=1, 1130 gap=0, 1131 scalarbar=True, 1132 # Figure and axes options: 1133 like=None, 1134 xlim=None, 1135 ylim=(None, None), 1136 zlim=(None, None), 1137 aspect=1, 1138 title="", 1139 xtitle=" ", 1140 ytitle=" ", 1141 ztitle="", 1142 ac="k", 1143 **fig_kwargs, 1144 ): 1145 """ 1146 Input data formats `[(x1,x2,..), (y1,y2,..)] or [(x1,y1), (x2,y2),..]` 1147 are both valid. 1148 1149 Use keyword `like=...` if you want to use the same format of a previously 1150 created Figure (useful when superimposing Figures) to make sure 1151 they are compatible and comparable. If they are not compatible 1152 you will receive an error message. 1153 1154 Arguments: 1155 bins : (list) 1156 binning as (nx, ny) 1157 weights : (list) 1158 array of weights to assign to each entry 1159 cmap : (str, lookuptable) 1160 color map name or look up table 1161 alpha : (float) 1162 opacity of the histogram 1163 gap : (float) 1164 separation between adjacent bins as a fraction for their size 1165 scalarbar : (bool) 1166 add a scalarbar to right of the histogram 1167 like : (Figure) 1168 grab and use the same format of the given Figure (for superimposing) 1169 xlim : (list) 1170 [x0, x1] range of interest. If left to None will automatically 1171 choose the minimum or the maximum of the data range. 1172 Data outside the range are completely ignored. 1173 ylim : (list) 1174 [y0, y1] range of interest. If left to None will automatically 1175 choose the minimum or the maximum of the data range. 1176 Data outside the range are completely ignored. 1177 aspect : (float) 1178 the desired aspect ratio of the figure. 1179 title : (str) 1180 title of the plot to appear on top. 1181 If left blank some statistics will be shown. 1182 xtitle : (str) 1183 x axis title 1184 ytitle : (str) 1185 y axis title 1186 ztitle : (str) 1187 title for the scalar bar 1188 ac : (str) 1189 axes color, additional keyword for Axes can also be added 1190 using e.g. `axes=dict(xygrid=True)` 1191 1192 Examples: 1193 - [histo_2d_a.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_2d_a.py) 1194 - [histo_2d_b.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_2d_b.py) 1195 1196  1197 """ 1198 xvalues = np.asarray(xvalues) 1199 if yvalues is None: 1200 # assume [(x1,y1), (x2,y2) ...] format 1201 yvalues = xvalues[:, 1] 1202 xvalues = xvalues[:, 0] 1203 else: 1204 yvalues = np.asarray(yvalues) 1205 1206 padding = [0, 0, 0, 0] 1207 1208 if like is None and vedo.last_figure is not None: 1209 if xlim is None and ylim == (None, None) and zlim == (None, None): 1210 like = vedo.last_figure 1211 1212 if like is not None: 1213 xlim = like.xlim 1214 ylim = like.ylim 1215 aspect = like.aspect 1216 padding = like.padding 1217 if bins is None: 1218 bins = like.bins 1219 if bins is None: 1220 bins = 20 1221 1222 if isinstance(bins, int): 1223 bins = (bins, bins) 1224 1225 if utils.is_sequence(xlim): 1226 # deal with user passing eg [x0, None] 1227 _x0, _x1 = xlim 1228 if _x0 is None: 1229 _x0 = xvalues.min() 1230 if _x1 is None: 1231 _x1 = xvalues.max() 1232 xlim = [_x0, _x1] 1233 1234 if utils.is_sequence(ylim): 1235 # deal with user passing eg [x0, None] 1236 _y0, _y1 = ylim 1237 if _y0 is None: 1238 _y0 = yvalues.min() 1239 if _y1 is None: 1240 _y1 = yvalues.max() 1241 ylim = [_y0, _y1] 1242 1243 H, xedges, yedges = np.histogram2d( 1244 xvalues, yvalues, weights=weights, bins=bins, range=(xlim, ylim) 1245 ) 1246 1247 xlim = np.min(xedges), np.max(xedges) 1248 ylim = np.min(yedges), np.max(yedges) 1249 dx, dy = xlim[1] - xlim[0], ylim[1] - ylim[0] 1250 1251 fig_kwargs["title"] = title 1252 fig_kwargs["xtitle"] = xtitle 1253 fig_kwargs["ytitle"] = ytitle 1254 fig_kwargs["ac"] = ac 1255 1256 self.entries = len(xvalues) 1257 self.frequencies = H 1258 self.edges = (xedges, yedges) 1259 self.mean = (xvalues.mean(), yvalues.mean()) 1260 self.std = (xvalues.std(), yvalues.std()) 1261 self.bins = bins # internally used by "like" 1262 1263 ############################### stats legend as htitle 1264 addstats = False 1265 if not title: 1266 if "axes" not in fig_kwargs: 1267 addstats = True 1268 axes_opts = {} 1269 fig_kwargs["axes"] = axes_opts 1270 elif fig_kwargs["axes"] is False: 1271 pass 1272 else: 1273 axes_opts = fig_kwargs["axes"] 1274 if "htitle" not in fig_kwargs["axes"]: 1275 addstats = True 1276 1277 if addstats: 1278 htitle = f"Entries:~~{int(self.entries)} " 1279 htitle += f"Mean:~~{utils.precision(self.mean, 3)} " 1280 htitle += f"STD:~~{utils.precision(self.std, 3)} " 1281 axes_opts["htitle"] = htitle 1282 axes_opts["htitle_justify"] = "bottom-left" 1283 axes_opts["htitle_size"] = 0.0175 1284 axes_opts["htitle_offset"] = [-0.49, 0.01, 0] 1285 1286 ############################################### Figure init 1287 Figure.__init__(self, xlim, ylim, aspect, padding, **fig_kwargs) 1288 1289 if self.yscale: 1290 ##################### the grid 1291 acts = [] 1292 g = shapes.Grid( 1293 pos=[(xlim[0] + xlim[1]) / 2, (ylim[0] + ylim[1]) / 2, 0], s=(dx, dy), res=bins[:2] 1294 ) 1295 g.alpha(alpha).lw(0).wireframe(False).flat().lighting("off") 1296 g.cmap(cmap, np.ravel(H.T), on="cells", vmin=zlim[0], vmax=zlim[1]) 1297 if gap: 1298 g.shrink(abs(1 - gap)) 1299 1300 if scalarbar: 1301 sc = g.add_scalarbar3d(ztitle, c=ac).scalarbar 1302 sc.scale([self.yscale, 1, 1]) ## prescale trick 1303 sbnds = sc.xbounds() 1304 sc.x(self.x1lim + (sbnds[1] - sbnds[0]) * 0.75) 1305 acts.append(sc) 1306 acts.append(g) 1307 1308 self.insert(*acts, as3d=False) 1309 self.name = "Histogram2D"
Input data formats [(x1,x2,..), (y1,y2,..)] or [(x1,y1), (x2,y2),..]
are both valid.
Use keyword like=... if you want to use the same format of a previously
created Figure (useful when superimposing Figures) to make sure
they are compatible and comparable. If they are not compatible
you will receive an error message.
Arguments:
- bins : (list) binning as (nx, ny)
- weights : (list) array of weights to assign to each entry
- cmap : (str, lookuptable) color map name or look up table
- alpha : (float) opacity of the histogram
- gap : (float) separation between adjacent bins as a fraction for their size
- scalarbar : (bool) add a scalarbar to right of the histogram
- like : (Figure) grab and use the same format of the given Figure (for superimposing)
- xlim : (list) [x0, x1] range of interest. If left to None will automatically choose the minimum or the maximum of the data range. Data outside the range are completely ignored.
- ylim : (list) [y0, y1] range of interest. If left to None will automatically choose the minimum or the maximum of the data range. Data outside the range are completely ignored.
- aspect : (float) the desired aspect ratio of the figure.
- title : (str) title of the plot to appear on top. If left blank some statistics will be shown.
- xtitle : (str) x axis title
- ytitle : (str) y axis title
- ztitle : (str) title for the scalar bar
- ac : (str)
axes color, additional keyword for Axes can also be added
using e.g.
axes=dict(xygrid=True)
Examples:
Inherited Members
1532class PlotXY(Figure): 1533 """Creates a `PlotXY(Figure)` object.""" 1534 1535 def __init__( 1536 self, 1537 # 1538 data, 1539 xerrors=None, 1540 yerrors=None, 1541 # 1542 lw=2, 1543 lc=None, 1544 la=1, 1545 dashed=False, 1546 splined=False, 1547 # 1548 elw=2, # error line width 1549 ec=None, # error line or band color 1550 error_band=False, # errors in x are ignored 1551 # 1552 marker="", 1553 ms=None, 1554 mc=None, 1555 ma=None, 1556 # Figure and axes options: 1557 like=None, 1558 xlim=None, 1559 ylim=(None, None), 1560 aspect=4 / 3, 1561 padding=0.05, 1562 # 1563 title="", 1564 xtitle=" ", 1565 ytitle=" ", 1566 ac="k", 1567 grid=True, 1568 ztolerance=None, 1569 label="", 1570 **fig_kwargs, 1571 ): 1572 """ 1573 Arguments: 1574 xerrors : (bool) 1575 show error bars associated to each point in x 1576 yerrors : (bool) 1577 show error bars associated to each point in y 1578 lw : (int) 1579 width of the line connecting points in pixel units. 1580 Set it to 0 to remove the line. 1581 lc : (str) 1582 line color 1583 la : (float) 1584 line "alpha", opacity of the line 1585 dashed : (bool) 1586 draw a dashed line instead of a continuous line 1587 splined : (bool) 1588 spline the line joining the point as a countinous curve 1589 elw : (int) 1590 width of error bar lines in units of pixels 1591 ec : (color) 1592 color of error bar, by default the same as marker color 1593 error_band : (bool) 1594 represent errors on y as a filled error band. 1595 Use `ec` keyword to modify its color. 1596 marker : (str, int) 1597 use a marker for the data points 1598 ms : (float) 1599 marker size 1600 mc : (color) 1601 color of the marker 1602 ma : (float) 1603 opacity of the marker 1604 xlim : (list) 1605 set limits to the range for the x variable 1606 ylim : (list) 1607 set limits to the range for the y variable 1608 aspect : (float, str) 1609 Desired aspect ratio. 1610 Use `aspect="equal"` to force the same units in x and y. 1611 Scaling factor is saved in Figure.yscale. 1612 padding : (float, list) 1613 keep a padding space from the axes (as a fraction of the axis size). 1614 This can be a list of four numbers. 1615 title : (str) 1616 title to appear on the top of the frame, like a header. 1617 xtitle : (str) 1618 title for the x-axis, can also be set using `axes=dict(xtitle="my x axis")` 1619 ytitle : (str) 1620 title for the y-axis, can also be set using `axes=dict(ytitle="my y axis")` 1621 ac : (str) 1622 axes color 1623 grid : (bool) 1624 show the background grid for the axes, can also be set using `axes=dict(xygrid=True)` 1625 ztolerance : (float) 1626 a tolerance factor to superimpose objects (along the z-axis). 1627 1628 Example: 1629 ```python 1630 import numpy as np 1631 from vedo.pyplot import plot 1632 x = np.arange(0, np.pi, 0.1) 1633 fig = plot(x, np.sin(2*x), 'r0-', aspect='equal') 1634 fig+= plot(x, np.cos(2*x), 'blue4 o-', like=fig) 1635 fig.show().close() 1636 ``` 1637  1638 1639 Examples: 1640 - [plot_errbars.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/plot_errbars.py) 1641 - [plot_errband.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/plot_errband.py) 1642 - [plot_pip.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/plot_pip.py) 1643 1644  1645 1646 - [scatter1.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/scatter1.py) 1647 - [scatter2.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/scatter2.py) 1648 1649  1650 """ 1651 line = False 1652 if lw > 0: 1653 line = True 1654 if marker == "" and not line and not splined: 1655 marker = "o" 1656 1657 if like is None and vedo.last_figure is not None: 1658 if xlim is None and ylim == (None, None): 1659 like = vedo.last_figure 1660 1661 if like is not None: 1662 xlim = like.xlim 1663 ylim = like.ylim 1664 aspect = like.aspect 1665 padding = like.padding 1666 1667 if utils.is_sequence(xlim): 1668 # deal with user passing eg [x0, None] 1669 _x0, _x1 = xlim 1670 if _x0 is None: 1671 _x0 = data.min() 1672 if _x1 is None: 1673 _x1 = data.max() 1674 xlim = [_x0, _x1] 1675 1676 # purge NaN from data 1677 validIds = np.all(np.logical_not(np.isnan(data))) 1678 data = np.array(data[validIds])[0] 1679 1680 fig_kwargs["title"] = title 1681 fig_kwargs["xtitle"] = xtitle 1682 fig_kwargs["ytitle"] = ytitle 1683 fig_kwargs["ac"] = ac 1684 fig_kwargs["ztolerance"] = ztolerance 1685 fig_kwargs["grid"] = grid 1686 1687 x0, y0 = np.min(data, axis=0) 1688 x1, y1 = np.max(data, axis=0) 1689 if xerrors is not None and not error_band: 1690 x0 = min(data[:, 0] - xerrors) 1691 x1 = max(data[:, 0] + xerrors) 1692 if yerrors is not None: 1693 y0 = min(data[:, 1] - yerrors) 1694 y1 = max(data[:, 1] + yerrors) 1695 1696 if like is None: 1697 if xlim is None: 1698 xlim = (None, None) 1699 xlim = list(xlim) 1700 if xlim[0] is None: 1701 xlim[0] = x0 1702 if xlim[1] is None: 1703 xlim[1] = x1 1704 ylim = list(ylim) 1705 if ylim[0] is None: 1706 ylim[0] = y0 1707 if ylim[1] is None: 1708 ylim[1] = y1 1709 1710 self.entries = len(data) 1711 self.mean = data.mean() 1712 self.std = data.std() 1713 1714 ######### the PlotXY marker 1715 # fall back solutions logic for colors 1716 if "c" in fig_kwargs: 1717 if mc is None: 1718 mc = fig_kwargs["c"] 1719 if lc is None: 1720 lc = fig_kwargs["c"] 1721 if ec is None: 1722 ec = fig_kwargs["c"] 1723 if lc is None: 1724 lc = "k" 1725 if mc is None: 1726 mc = lc 1727 if ma is None: 1728 ma = la 1729 if ec is None: 1730 if mc is None: 1731 ec = lc 1732 else: 1733 ec = mc 1734 1735 if label: 1736 nlab = LabelData() 1737 nlab.text = label 1738 nlab.tcolor = ac 1739 nlab.marker = marker 1740 if line and marker == "": 1741 nlab.marker = "-" 1742 nlab.mcolor = mc 1743 fig_kwargs["label"] = nlab 1744 1745 ############################################### Figure init 1746 Figure.__init__(self, xlim, ylim, aspect, padding, **fig_kwargs) 1747 1748 if not self.yscale: 1749 return 1750 1751 acts = [] 1752 1753 ######### the PlotXY Line or Spline 1754 if dashed: 1755 l = shapes.DashedLine(data, c=lc, alpha=la, lw=lw) 1756 acts.append(l) 1757 elif splined: 1758 l = shapes.KSpline(data).lw(lw).c(lc).alpha(la) 1759 acts.append(l) 1760 elif line: 1761 l = shapes.Line(data, c=lc, alpha=la).lw(lw) 1762 acts.append(l) 1763 1764 if marker: 1765 1766 pts = np.c_[data, np.zeros(len(data))] 1767 1768 if utils.is_sequence(ms): 1769 ### variable point size 1770 mk = shapes.Marker(marker, s=1) 1771 mk.scale([1, 1 / self.yscale, 1]) 1772 msv = np.zeros_like(pts) 1773 msv[:, 0] = ms 1774 marked = shapes.Glyph( 1775 pts, mk, c=mc, orientation_array=msv, scale_by_vector_size=True 1776 ) 1777 else: 1778 ### fixed point size 1779 if ms is None: 1780 ms = (xlim[1] - xlim[0]) / 100.0 1781 1782 if utils.is_sequence(mc): 1783 fig_kwargs["marker_color"] = None # for labels 1784 mk = shapes.Marker(marker, s=ms) 1785 mk.scale([1, 1 / self.yscale, 1]) 1786 msv = np.zeros_like(pts) 1787 msv[:, 0] = 1 1788 marked = shapes.Glyph( 1789 pts, mk, c=mc, orientation_array=msv, scale_by_vector_size=True 1790 ) 1791 else: 1792 mk = shapes.Marker(marker, s=ms) 1793 mk.scale([1, 1 / self.yscale, 1]) 1794 marked = shapes.Glyph(pts, mk, c=mc) 1795 1796 marked.name = "Marker" 1797 marked.alpha(ma) 1798 marked.z(3 * self.ztolerance) 1799 acts.append(marked) 1800 1801 ######### the PlotXY marker errors 1802 ztol = self.ztolerance 1803 1804 if error_band: 1805 yerrors = np.abs(yerrors) 1806 du = np.array(data) 1807 dd = np.array(data) 1808 du[:, 1] += yerrors 1809 dd[:, 1] -= yerrors 1810 if splined: 1811 res = len(data) * 20 1812 band1 = shapes.KSpline(du, res=res) 1813 band2 = shapes.KSpline(dd, res=res) 1814 band = shapes.Ribbon(band1, band2, res=(res, 2)) 1815 else: 1816 dd = list(reversed(dd.tolist())) 1817 band = shapes.Line(du.tolist() + dd, closed=True) 1818 band.triangulate().lw(0) 1819 if ec is None: 1820 band.c(lc) 1821 else: 1822 band.c(ec) 1823 band.lighting("off").alpha(la).z(ztol / 20) 1824 acts.append(band) 1825 1826 else: 1827 1828 ## xerrors 1829 if xerrors is not None: 1830 if len(xerrors) == len(data): 1831 errs = [] 1832 for i, val in enumerate(data): 1833 xval, yval = val 1834 xerr = xerrors[i] / 2 1835 el = shapes.Line((xval - xerr, yval, ztol), (xval + xerr, yval, ztol)) 1836 el.lw(elw) 1837 errs.append(el) 1838 mxerrs = merge(errs).c(ec).lw(lw).alpha(ma).z(2 * ztol) 1839 acts.append(mxerrs) 1840 else: 1841 vedo.logger.error("in PlotXY(xerrors=...): mismatch in array length") 1842 1843 ## yerrors 1844 if yerrors is not None: 1845 if len(yerrors) == len(data): 1846 errs = [] 1847 for i, val in enumerate(data): 1848 xval, yval = val 1849 yerr = yerrors[i] 1850 el = shapes.Line((xval, yval - yerr, ztol), (xval, yval + yerr, ztol)) 1851 el.lw(elw) 1852 errs.append(el) 1853 myerrs = merge(errs).c(ec).lw(lw).alpha(ma).z(2 * ztol) 1854 acts.append(myerrs) 1855 else: 1856 vedo.logger.error("in PlotXY(yerrors=...): mismatch in array length") 1857 1858 self.insert(*acts, as3d=False) 1859 self.name = "PlotXY"
Creates a PlotXY(Figure) object.
1535 def __init__( 1536 self, 1537 # 1538 data, 1539 xerrors=None, 1540 yerrors=None, 1541 # 1542 lw=2, 1543 lc=None, 1544 la=1, 1545 dashed=False, 1546 splined=False, 1547 # 1548 elw=2, # error line width 1549 ec=None, # error line or band color 1550 error_band=False, # errors in x are ignored 1551 # 1552 marker="", 1553 ms=None, 1554 mc=None, 1555 ma=None, 1556 # Figure and axes options: 1557 like=None, 1558 xlim=None, 1559 ylim=(None, None), 1560 aspect=4 / 3, 1561 padding=0.05, 1562 # 1563 title="", 1564 xtitle=" ", 1565 ytitle=" ", 1566 ac="k", 1567 grid=True, 1568 ztolerance=None, 1569 label="", 1570 **fig_kwargs, 1571 ): 1572 """ 1573 Arguments: 1574 xerrors : (bool) 1575 show error bars associated to each point in x 1576 yerrors : (bool) 1577 show error bars associated to each point in y 1578 lw : (int) 1579 width of the line connecting points in pixel units. 1580 Set it to 0 to remove the line. 1581 lc : (str) 1582 line color 1583 la : (float) 1584 line "alpha", opacity of the line 1585 dashed : (bool) 1586 draw a dashed line instead of a continuous line 1587 splined : (bool) 1588 spline the line joining the point as a countinous curve 1589 elw : (int) 1590 width of error bar lines in units of pixels 1591 ec : (color) 1592 color of error bar, by default the same as marker color 1593 error_band : (bool) 1594 represent errors on y as a filled error band. 1595 Use `ec` keyword to modify its color. 1596 marker : (str, int) 1597 use a marker for the data points 1598 ms : (float) 1599 marker size 1600 mc : (color) 1601 color of the marker 1602 ma : (float) 1603 opacity of the marker 1604 xlim : (list) 1605 set limits to the range for the x variable 1606 ylim : (list) 1607 set limits to the range for the y variable 1608 aspect : (float, str) 1609 Desired aspect ratio. 1610 Use `aspect="equal"` to force the same units in x and y. 1611 Scaling factor is saved in Figure.yscale. 1612 padding : (float, list) 1613 keep a padding space from the axes (as a fraction of the axis size). 1614 This can be a list of four numbers. 1615 title : (str) 1616 title to appear on the top of the frame, like a header. 1617 xtitle : (str) 1618 title for the x-axis, can also be set using `axes=dict(xtitle="my x axis")` 1619 ytitle : (str) 1620 title for the y-axis, can also be set using `axes=dict(ytitle="my y axis")` 1621 ac : (str) 1622 axes color 1623 grid : (bool) 1624 show the background grid for the axes, can also be set using `axes=dict(xygrid=True)` 1625 ztolerance : (float) 1626 a tolerance factor to superimpose objects (along the z-axis). 1627 1628 Example: 1629 ```python 1630 import numpy as np 1631 from vedo.pyplot import plot 1632 x = np.arange(0, np.pi, 0.1) 1633 fig = plot(x, np.sin(2*x), 'r0-', aspect='equal') 1634 fig+= plot(x, np.cos(2*x), 'blue4 o-', like=fig) 1635 fig.show().close() 1636 ``` 1637  1638 1639 Examples: 1640 - [plot_errbars.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/plot_errbars.py) 1641 - [plot_errband.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/plot_errband.py) 1642 - [plot_pip.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/plot_pip.py) 1643 1644  1645 1646 - [scatter1.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/scatter1.py) 1647 - [scatter2.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/scatter2.py) 1648 1649  1650 """ 1651 line = False 1652 if lw > 0: 1653 line = True 1654 if marker == "" and not line and not splined: 1655 marker = "o" 1656 1657 if like is None and vedo.last_figure is not None: 1658 if xlim is None and ylim == (None, None): 1659 like = vedo.last_figure 1660 1661 if like is not None: 1662 xlim = like.xlim 1663 ylim = like.ylim 1664 aspect = like.aspect 1665 padding = like.padding 1666 1667 if utils.is_sequence(xlim): 1668 # deal with user passing eg [x0, None] 1669 _x0, _x1 = xlim 1670 if _x0 is None: 1671 _x0 = data.min() 1672 if _x1 is None: 1673 _x1 = data.max() 1674 xlim = [_x0, _x1] 1675 1676 # purge NaN from data 1677 validIds = np.all(np.logical_not(np.isnan(data))) 1678 data = np.array(data[validIds])[0] 1679 1680 fig_kwargs["title"] = title 1681 fig_kwargs["xtitle"] = xtitle 1682 fig_kwargs["ytitle"] = ytitle 1683 fig_kwargs["ac"] = ac 1684 fig_kwargs["ztolerance"] = ztolerance 1685 fig_kwargs["grid"] = grid 1686 1687 x0, y0 = np.min(data, axis=0) 1688 x1, y1 = np.max(data, axis=0) 1689 if xerrors is not None and not error_band: 1690 x0 = min(data[:, 0] - xerrors) 1691 x1 = max(data[:, 0] + xerrors) 1692 if yerrors is not None: 1693 y0 = min(data[:, 1] - yerrors) 1694 y1 = max(data[:, 1] + yerrors) 1695 1696 if like is None: 1697 if xlim is None: 1698 xlim = (None, None) 1699 xlim = list(xlim) 1700 if xlim[0] is None: 1701 xlim[0] = x0 1702 if xlim[1] is None: 1703 xlim[1] = x1 1704 ylim = list(ylim) 1705 if ylim[0] is None: 1706 ylim[0] = y0 1707 if ylim[1] is None: 1708 ylim[1] = y1 1709 1710 self.entries = len(data) 1711 self.mean = data.mean() 1712 self.std = data.std() 1713 1714 ######### the PlotXY marker 1715 # fall back solutions logic for colors 1716 if "c" in fig_kwargs: 1717 if mc is None: 1718 mc = fig_kwargs["c"] 1719 if lc is None: 1720 lc = fig_kwargs["c"] 1721 if ec is None: 1722 ec = fig_kwargs["c"] 1723 if lc is None: 1724 lc = "k" 1725 if mc is None: 1726 mc = lc 1727 if ma is None: 1728 ma = la 1729 if ec is None: 1730 if mc is None: 1731 ec = lc 1732 else: 1733 ec = mc 1734 1735 if label: 1736 nlab = LabelData() 1737 nlab.text = label 1738 nlab.tcolor = ac 1739 nlab.marker = marker 1740 if line and marker == "": 1741 nlab.marker = "-" 1742 nlab.mcolor = mc 1743 fig_kwargs["label"] = nlab 1744 1745 ############################################### Figure init 1746 Figure.__init__(self, xlim, ylim, aspect, padding, **fig_kwargs) 1747 1748 if not self.yscale: 1749 return 1750 1751 acts = [] 1752 1753 ######### the PlotXY Line or Spline 1754 if dashed: 1755 l = shapes.DashedLine(data, c=lc, alpha=la, lw=lw) 1756 acts.append(l) 1757 elif splined: 1758 l = shapes.KSpline(data).lw(lw).c(lc).alpha(la) 1759 acts.append(l) 1760 elif line: 1761 l = shapes.Line(data, c=lc, alpha=la).lw(lw) 1762 acts.append(l) 1763 1764 if marker: 1765 1766 pts = np.c_[data, np.zeros(len(data))] 1767 1768 if utils.is_sequence(ms): 1769 ### variable point size 1770 mk = shapes.Marker(marker, s=1) 1771 mk.scale([1, 1 / self.yscale, 1]) 1772 msv = np.zeros_like(pts) 1773 msv[:, 0] = ms 1774 marked = shapes.Glyph( 1775 pts, mk, c=mc, orientation_array=msv, scale_by_vector_size=True 1776 ) 1777 else: 1778 ### fixed point size 1779 if ms is None: 1780 ms = (xlim[1] - xlim[0]) / 100.0 1781 1782 if utils.is_sequence(mc): 1783 fig_kwargs["marker_color"] = None # for labels 1784 mk = shapes.Marker(marker, s=ms) 1785 mk.scale([1, 1 / self.yscale, 1]) 1786 msv = np.zeros_like(pts) 1787 msv[:, 0] = 1 1788 marked = shapes.Glyph( 1789 pts, mk, c=mc, orientation_array=msv, scale_by_vector_size=True 1790 ) 1791 else: 1792 mk = shapes.Marker(marker, s=ms) 1793 mk.scale([1, 1 / self.yscale, 1]) 1794 marked = shapes.Glyph(pts, mk, c=mc) 1795 1796 marked.name = "Marker" 1797 marked.alpha(ma) 1798 marked.z(3 * self.ztolerance) 1799 acts.append(marked) 1800 1801 ######### the PlotXY marker errors 1802 ztol = self.ztolerance 1803 1804 if error_band: 1805 yerrors = np.abs(yerrors) 1806 du = np.array(data) 1807 dd = np.array(data) 1808 du[:, 1] += yerrors 1809 dd[:, 1] -= yerrors 1810 if splined: 1811 res = len(data) * 20 1812 band1 = shapes.KSpline(du, res=res) 1813 band2 = shapes.KSpline(dd, res=res) 1814 band = shapes.Ribbon(band1, band2, res=(res, 2)) 1815 else: 1816 dd = list(reversed(dd.tolist())) 1817 band = shapes.Line(du.tolist() + dd, closed=True) 1818 band.triangulate().lw(0) 1819 if ec is None: 1820 band.c(lc) 1821 else: 1822 band.c(ec) 1823 band.lighting("off").alpha(la).z(ztol / 20) 1824 acts.append(band) 1825 1826 else: 1827 1828 ## xerrors 1829 if xerrors is not None: 1830 if len(xerrors) == len(data): 1831 errs = [] 1832 for i, val in enumerate(data): 1833 xval, yval = val 1834 xerr = xerrors[i] / 2 1835 el = shapes.Line((xval - xerr, yval, ztol), (xval + xerr, yval, ztol)) 1836 el.lw(elw) 1837 errs.append(el) 1838 mxerrs = merge(errs).c(ec).lw(lw).alpha(ma).z(2 * ztol) 1839 acts.append(mxerrs) 1840 else: 1841 vedo.logger.error("in PlotXY(xerrors=...): mismatch in array length") 1842 1843 ## yerrors 1844 if yerrors is not None: 1845 if len(yerrors) == len(data): 1846 errs = [] 1847 for i, val in enumerate(data): 1848 xval, yval = val 1849 yerr = yerrors[i] 1850 el = shapes.Line((xval, yval - yerr, ztol), (xval, yval + yerr, ztol)) 1851 el.lw(elw) 1852 errs.append(el) 1853 myerrs = merge(errs).c(ec).lw(lw).alpha(ma).z(2 * ztol) 1854 acts.append(myerrs) 1855 else: 1856 vedo.logger.error("in PlotXY(yerrors=...): mismatch in array length") 1857 1858 self.insert(*acts, as3d=False) 1859 self.name = "PlotXY"
Arguments:
- xerrors : (bool) show error bars associated to each point in x
- yerrors : (bool) show error bars associated to each point in y
- lw : (int) width of the line connecting points in pixel units. Set it to 0 to remove the line.
- lc : (str) line color
- la : (float) line "alpha", opacity of the line
- dashed : (bool) draw a dashed line instead of a continuous line
- splined : (bool) spline the line joining the point as a countinous curve
- elw : (int) width of error bar lines in units of pixels
- ec : (color) color of error bar, by default the same as marker color
- error_band : (bool)
represent errors on y as a filled error band.
Use
eckeyword to modify its color. - marker : (str, int) use a marker for the data points
- ms : (float) marker size
- mc : (color) color of the marker
- ma : (float) opacity of the marker
- xlim : (list) set limits to the range for the x variable
- ylim : (list) set limits to the range for the y variable
- aspect : (float, str)
Desired aspect ratio.
Use
aspect="equal"to force the same units in x and y. Scaling factor is saved in Figure.yscale. - padding : (float, list) keep a padding space from the axes (as a fraction of the axis size). This can be a list of four numbers.
- title : (str) title to appear on the top of the frame, like a header.
- xtitle : (str)
title for the x-axis, can also be set using
axes=dict(xtitle="my x axis") - ytitle : (str)
title for the y-axis, can also be set using
axes=dict(ytitle="my y axis") - ac : (str) axes color
- grid : (bool)
show the background grid for the axes, can also be set using
axes=dict(xygrid=True) - ztolerance : (float) a tolerance factor to superimpose objects (along the z-axis).
Example:
import numpy as np from vedo.pyplot import plot x = np.arange(0, np.pi, 0.1) fig = plot(x, np.sin(2*x), 'r0-', aspect='equal') fig+= plot(x, np.cos(2*x), 'blue4 o-', like=fig) fig.show().close()
Examples:
Inherited Members
1313class PlotBars(Figure): 1314 """Creates a `PlotBars(Figure)` object.""" 1315 1316 def __init__( 1317 self, 1318 data, 1319 errors=False, 1320 logscale=False, 1321 fill=True, 1322 gap=0.02, 1323 radius=0.05, 1324 c="olivedrab", 1325 alpha=1, 1326 texture="", 1327 outline=False, 1328 lw=2, 1329 lc="k", 1330 # Figure and axes options: 1331 like=None, 1332 xlim=(None, None), 1333 ylim=(0, None), 1334 aspect=4 / 3, 1335 padding=(0.025, 0.025, 0, 0.05), 1336 # 1337 title="", 1338 xtitle=" ", 1339 ytitle=" ", 1340 ac="k", 1341 grid=False, 1342 ztolerance=None, 1343 **fig_kwargs, 1344 ): 1345 """ 1346 Input must be in format `[counts, labels, colors, edges]`. 1347 Either or both `edges` and `colors` are optional and can be omitted. 1348 1349 Use keyword `like=...` if you want to use the same format of a previously 1350 created Figure (useful when superimposing Figures) to make sure 1351 they are compatible and comparable. If they are not compatible 1352 you will receive an error message. 1353 1354 Arguments: 1355 errors : (bool) 1356 show error bars 1357 logscale : (bool) 1358 use logscale on y-axis 1359 fill : (bool) 1360 fill bars with solid color `c` 1361 gap : (float) 1362 leave a small space btw bars 1363 radius : (float) 1364 border radius of the top of the histogram bar. Default value is 0.1. 1365 texture : (str) 1366 url or path to an image to be used as texture for the bin 1367 outline : (bool) 1368 show outline of the bins 1369 xtitle : (str) 1370 title for the x-axis, can also be set using `axes=dict(xtitle="my x axis")` 1371 ytitle : (str) 1372 title for the y-axis, can also be set using `axes=dict(ytitle="my y axis")` 1373 ac : (str) 1374 axes color 1375 padding : (float, list) 1376 keep a padding space from the axes (as a fraction of the axis size). 1377 This can be a list of four numbers. 1378 aspect : (float) 1379 the desired aspect ratio of the figure. Default is 4/3. 1380 grid : (bool) 1381 show the background grid for the axes, can also be set using `axes=dict(xygrid=True)` 1382 1383 Examples: 1384 - [plot_bars.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/plot_bars.py) 1385 1386  1387 """ 1388 ndata = len(data) 1389 if ndata == 4: 1390 counts, xlabs, cols, edges = data 1391 elif ndata == 3: 1392 counts, xlabs, cols = data 1393 edges = np.array(range(len(counts) + 1)) + 0.5 1394 elif ndata == 2: 1395 counts, xlabs = data 1396 edges = np.array(range(len(counts) + 1)) + 0.5 1397 cols = [c] * len(counts) 1398 else: 1399 m = "barplot error: data must be given as [counts, labels, colors, edges] not\n" 1400 vedo.logger.error(m + f" {data}\n bin edges and colors are optional.") 1401 raise RuntimeError() 1402 1403 # sanity checks 1404 assert len(counts) == len(xlabs) 1405 assert len(counts) == len(cols) 1406 assert len(counts) == len(edges) - 1 1407 1408 counts = np.asarray(counts) 1409 edges = np.asarray(edges) 1410 1411 if logscale: 1412 counts = np.log10(counts + 1) 1413 if ytitle == " ": 1414 ytitle = "log_10 (counts+1)" 1415 1416 if like is None and vedo.last_figure is not None: 1417 if xlim == (None, None) and ylim == (0, None): 1418 like = vedo.last_figure 1419 1420 if like is not None: 1421 xlim = like.xlim 1422 ylim = like.ylim 1423 aspect = like.aspect 1424 padding = like.padding 1425 1426 if utils.is_sequence(xlim): 1427 # deal with user passing eg [x0, None] 1428 _x0, _x1 = xlim 1429 if _x0 is None: 1430 _x0 = np.min(edges) 1431 if _x1 is None: 1432 _x1 = np.max(edges) 1433 xlim = [_x0, _x1] 1434 1435 x0, x1 = np.min(edges), np.max(edges) 1436 y0, y1 = ylim[0], np.max(counts) 1437 1438 if like is None: 1439 ylim = list(ylim) 1440 if xlim is None: 1441 xlim = [x0, x1] 1442 if ylim[1] is None: 1443 ylim[1] = y1 1444 if ylim[0] != 0: 1445 ylim[0] = y0 1446 1447 fig_kwargs["title"] = title 1448 fig_kwargs["xtitle"] = xtitle 1449 fig_kwargs["ytitle"] = ytitle 1450 fig_kwargs["ac"] = ac 1451 fig_kwargs["ztolerance"] = ztolerance 1452 fig_kwargs["grid"] = grid 1453 1454 centers = (edges[0:-1] + edges[1:]) / 2 1455 binsizes = (centers - edges[0:-1]) * 2 1456 1457 if "axes" not in fig_kwargs: 1458 fig_kwargs["axes"] = {} 1459 1460 _xlabs = [] 1461 for center, xlb in zip(centers, xlabs): 1462 _xlabs.append([center, str(xlb)]) 1463 fig_kwargs["axes"]["x_values_and_labels"] = _xlabs 1464 1465 ############################################### Figure 1466 self.statslegend = "" 1467 self.edges = edges 1468 self.centers = centers 1469 self.bins = edges # internal used by "like" 1470 Figure.__init__(self, xlim, ylim, aspect, padding, **fig_kwargs) 1471 if not self.yscale: 1472 return 1473 1474 rs = [] 1475 maxheigth = 0 1476 if fill: ##################### 1477 if outline: 1478 gap = 0 1479 1480 for i in range(len(centers)): 1481 binsize = binsizes[i] 1482 p0 = (edges[i] + gap * binsize, 0, 0) 1483 p1 = (edges[i + 1] - gap * binsize, counts[i], 0) 1484 1485 if radius: 1486 rds = np.array([0, 0, radius, radius]) 1487 p1_yscaled = [p1[0], p1[1] * self.yscale, 0] 1488 r = shapes.Rectangle(p0, p1_yscaled, radius=rds * binsize, res=6) 1489 r.scale([1, 1 / self.yscale, 1]) 1490 r.radius = None # so it doesnt get recreated and rescaled by insert() 1491 else: 1492 r = shapes.Rectangle(p0, p1) 1493 1494 if texture: 1495 r.texture(texture) 1496 c = "w" 1497 1498 r.PickableOff() 1499 maxheigth = max(maxheigth, p1[1]) 1500 if c in colors.cmaps_names: 1501 col = colors.color_map((p0[0] + p1[0]) / 2, c, edges[0], edges[-1]) 1502 else: 1503 col = cols[i] 1504 r.color(col).alpha(alpha).lighting("off") 1505 r.name = f"bar_{i}" 1506 r.z(self.ztolerance) 1507 rs.append(r) 1508 1509 elif outline: ##################### 1510 lns = [[edges[0], 0, 0]] 1511 for i in range(len(centers)): 1512 lns.append([edges[i], counts[i], 0]) 1513 lns.append([edges[i + 1], counts[i], 0]) 1514 maxheigth = max(maxheigth, counts[i]) 1515 lns.append([edges[-1], 0, 0]) 1516 outl = shapes.Line(lns, c=lc, alpha=alpha, lw=lw).z(self.ztolerance) 1517 outl.name = f"bar_outline_{i}" 1518 rs.append(outl) 1519 1520 if errors: ##################### 1521 for x, f in centers: 1522 err = np.sqrt(f) 1523 el = shapes.Line([x, f - err / 2, 0], [x, f + err / 2, 0], c=lc, alpha=alpha, lw=lw) 1524 el.z(self.ztolerance * 2) 1525 rs.append(el) 1526 1527 self.insert(*rs, as3d=False) 1528 self.name = "PlotBars"
Creates a PlotBars(Figure) object.
1316 def __init__( 1317 self, 1318 data, 1319 errors=False, 1320 logscale=False, 1321 fill=True, 1322 gap=0.02, 1323 radius=0.05, 1324 c="olivedrab", 1325 alpha=1, 1326 texture="", 1327 outline=False, 1328 lw=2, 1329 lc="k", 1330 # Figure and axes options: 1331 like=None, 1332 xlim=(None, None), 1333 ylim=(0, None), 1334 aspect=4 / 3, 1335 padding=(0.025, 0.025, 0, 0.05), 1336 # 1337 title="", 1338 xtitle=" ", 1339 ytitle=" ", 1340 ac="k", 1341 grid=False, 1342 ztolerance=None, 1343 **fig_kwargs, 1344 ): 1345 """ 1346 Input must be in format `[counts, labels, colors, edges]`. 1347 Either or both `edges` and `colors` are optional and can be omitted. 1348 1349 Use keyword `like=...` if you want to use the same format of a previously 1350 created Figure (useful when superimposing Figures) to make sure 1351 they are compatible and comparable. If they are not compatible 1352 you will receive an error message. 1353 1354 Arguments: 1355 errors : (bool) 1356 show error bars 1357 logscale : (bool) 1358 use logscale on y-axis 1359 fill : (bool) 1360 fill bars with solid color `c` 1361 gap : (float) 1362 leave a small space btw bars 1363 radius : (float) 1364 border radius of the top of the histogram bar. Default value is 0.1. 1365 texture : (str) 1366 url or path to an image to be used as texture for the bin 1367 outline : (bool) 1368 show outline of the bins 1369 xtitle : (str) 1370 title for the x-axis, can also be set using `axes=dict(xtitle="my x axis")` 1371 ytitle : (str) 1372 title for the y-axis, can also be set using `axes=dict(ytitle="my y axis")` 1373 ac : (str) 1374 axes color 1375 padding : (float, list) 1376 keep a padding space from the axes (as a fraction of the axis size). 1377 This can be a list of four numbers. 1378 aspect : (float) 1379 the desired aspect ratio of the figure. Default is 4/3. 1380 grid : (bool) 1381 show the background grid for the axes, can also be set using `axes=dict(xygrid=True)` 1382 1383 Examples: 1384 - [plot_bars.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/plot_bars.py) 1385 1386  1387 """ 1388 ndata = len(data) 1389 if ndata == 4: 1390 counts, xlabs, cols, edges = data 1391 elif ndata == 3: 1392 counts, xlabs, cols = data 1393 edges = np.array(range(len(counts) + 1)) + 0.5 1394 elif ndata == 2: 1395 counts, xlabs = data 1396 edges = np.array(range(len(counts) + 1)) + 0.5 1397 cols = [c] * len(counts) 1398 else: 1399 m = "barplot error: data must be given as [counts, labels, colors, edges] not\n" 1400 vedo.logger.error(m + f" {data}\n bin edges and colors are optional.") 1401 raise RuntimeError() 1402 1403 # sanity checks 1404 assert len(counts) == len(xlabs) 1405 assert len(counts) == len(cols) 1406 assert len(counts) == len(edges) - 1 1407 1408 counts = np.asarray(counts) 1409 edges = np.asarray(edges) 1410 1411 if logscale: 1412 counts = np.log10(counts + 1) 1413 if ytitle == " ": 1414 ytitle = "log_10 (counts+1)" 1415 1416 if like is None and vedo.last_figure is not None: 1417 if xlim == (None, None) and ylim == (0, None): 1418 like = vedo.last_figure 1419 1420 if like is not None: 1421 xlim = like.xlim 1422 ylim = like.ylim 1423 aspect = like.aspect 1424 padding = like.padding 1425 1426 if utils.is_sequence(xlim): 1427 # deal with user passing eg [x0, None] 1428 _x0, _x1 = xlim 1429 if _x0 is None: 1430 _x0 = np.min(edges) 1431 if _x1 is None: 1432 _x1 = np.max(edges) 1433 xlim = [_x0, _x1] 1434 1435 x0, x1 = np.min(edges), np.max(edges) 1436 y0, y1 = ylim[0], np.max(counts) 1437 1438 if like is None: 1439 ylim = list(ylim) 1440 if xlim is None: 1441 xlim = [x0, x1] 1442 if ylim[1] is None: 1443 ylim[1] = y1 1444 if ylim[0] != 0: 1445 ylim[0] = y0 1446 1447 fig_kwargs["title"] = title 1448 fig_kwargs["xtitle"] = xtitle 1449 fig_kwargs["ytitle"] = ytitle 1450 fig_kwargs["ac"] = ac 1451 fig_kwargs["ztolerance"] = ztolerance 1452 fig_kwargs["grid"] = grid 1453 1454 centers = (edges[0:-1] + edges[1:]) / 2 1455 binsizes = (centers - edges[0:-1]) * 2 1456 1457 if "axes" not in fig_kwargs: 1458 fig_kwargs["axes"] = {} 1459 1460 _xlabs = [] 1461 for center, xlb in zip(centers, xlabs): 1462 _xlabs.append([center, str(xlb)]) 1463 fig_kwargs["axes"]["x_values_and_labels"] = _xlabs 1464 1465 ############################################### Figure 1466 self.statslegend = "" 1467 self.edges = edges 1468 self.centers = centers 1469 self.bins = edges # internal used by "like" 1470 Figure.__init__(self, xlim, ylim, aspect, padding, **fig_kwargs) 1471 if not self.yscale: 1472 return 1473 1474 rs = [] 1475 maxheigth = 0 1476 if fill: ##################### 1477 if outline: 1478 gap = 0 1479 1480 for i in range(len(centers)): 1481 binsize = binsizes[i] 1482 p0 = (edges[i] + gap * binsize, 0, 0) 1483 p1 = (edges[i + 1] - gap * binsize, counts[i], 0) 1484 1485 if radius: 1486 rds = np.array([0, 0, radius, radius]) 1487 p1_yscaled = [p1[0], p1[1] * self.yscale, 0] 1488 r = shapes.Rectangle(p0, p1_yscaled, radius=rds * binsize, res=6) 1489 r.scale([1, 1 / self.yscale, 1]) 1490 r.radius = None # so it doesnt get recreated and rescaled by insert() 1491 else: 1492 r = shapes.Rectangle(p0, p1) 1493 1494 if texture: 1495 r.texture(texture) 1496 c = "w" 1497 1498 r.PickableOff() 1499 maxheigth = max(maxheigth, p1[1]) 1500 if c in colors.cmaps_names: 1501 col = colors.color_map((p0[0] + p1[0]) / 2, c, edges[0], edges[-1]) 1502 else: 1503 col = cols[i] 1504 r.color(col).alpha(alpha).lighting("off") 1505 r.name = f"bar_{i}" 1506 r.z(self.ztolerance) 1507 rs.append(r) 1508 1509 elif outline: ##################### 1510 lns = [[edges[0], 0, 0]] 1511 for i in range(len(centers)): 1512 lns.append([edges[i], counts[i], 0]) 1513 lns.append([edges[i + 1], counts[i], 0]) 1514 maxheigth = max(maxheigth, counts[i]) 1515 lns.append([edges[-1], 0, 0]) 1516 outl = shapes.Line(lns, c=lc, alpha=alpha, lw=lw).z(self.ztolerance) 1517 outl.name = f"bar_outline_{i}" 1518 rs.append(outl) 1519 1520 if errors: ##################### 1521 for x, f in centers: 1522 err = np.sqrt(f) 1523 el = shapes.Line([x, f - err / 2, 0], [x, f + err / 2, 0], c=lc, alpha=alpha, lw=lw) 1524 el.z(self.ztolerance * 2) 1525 rs.append(el) 1526 1527 self.insert(*rs, as3d=False) 1528 self.name = "PlotBars"
Input must be in format [counts, labels, colors, edges].
Either or both edges and colors are optional and can be omitted.
Use keyword like=... if you want to use the same format of a previously
created Figure (useful when superimposing Figures) to make sure
they are compatible and comparable. If they are not compatible
you will receive an error message.
Arguments:
- errors : (bool) show error bars
- logscale : (bool) use logscale on y-axis
- fill : (bool)
fill bars with solid color
c - gap : (float) leave a small space btw bars
- radius : (float) border radius of the top of the histogram bar. Default value is 0.1.
- texture : (str) url or path to an image to be used as texture for the bin
- outline : (bool) show outline of the bins
- xtitle : (str)
title for the x-axis, can also be set using
axes=dict(xtitle="my x axis") - ytitle : (str)
title for the y-axis, can also be set using
axes=dict(ytitle="my y axis") - ac : (str) axes color
- padding : (float, list) keep a padding space from the axes (as a fraction of the axis size). This can be a list of four numbers.
- aspect : (float) the desired aspect ratio of the figure. Default is 4/3.
- grid : (bool)
show the background grid for the axes, can also be set using
axes=dict(xygrid=True)
Examples:
Inherited Members
1862def plot(*args, **kwargs): 1863 """ 1864 Draw a 2D line plot, or scatter plot, of variable x vs variable y. 1865 Input format can be either `[allx], [allx, ally] or [(x1,y1), (x2,y2), ...]` 1866 1867 Use `like=...` if you want to use the same format of a previously 1868 created Figure (useful when superimposing Figures) to make sure 1869 they are compatible and comparable. If they are not compatible 1870 you will receive an error message. 1871 1872 Arguments: 1873 xerrors : (bool) 1874 show error bars associated to each point in x 1875 yerrors : (bool) 1876 show error bars associated to each point in y 1877 lw : (int) 1878 width of the line connecting points in pixel units. 1879 Set it to 0 to remove the line. 1880 lc : (str) 1881 line color 1882 la : (float) 1883 line "alpha", opacity of the line 1884 dashed : (bool) 1885 draw a dashed line instead of a continuous line 1886 splined : (bool) 1887 spline the line joining the point as a countinous curve 1888 elw : (int) 1889 width of error bar lines in units of pixels 1890 ec : (color) 1891 color of error bar, by default the same as marker color 1892 error_band : (bool) 1893 represent errors on y as a filled error band. 1894 Use `ec` keyword to modify its color. 1895 marker : (str, int) 1896 use a marker for the data points 1897 ms : (float) 1898 marker size 1899 mc : (color) 1900 color of the marker 1901 ma : (float) 1902 opacity of the marker 1903 xlim : (list) 1904 set limits to the range for the x variable 1905 ylim : (list) 1906 set limits to the range for the y variable 1907 aspect : (float) 1908 Desired aspect ratio. 1909 If None, it is automatically calculated to get a reasonable aspect ratio. 1910 Scaling factor is saved in Figure.yscale 1911 padding : (float, list) 1912 keep a padding space from the axes (as a fraction of the axis size). 1913 This can be a list of four numbers. 1914 title : (str) 1915 title to appear on the top of the frame, like a header. 1916 xtitle : (str) 1917 title for the x-axis, can also be set using `axes=dict(xtitle="my x axis")` 1918 ytitle : (str) 1919 title for the y-axis, can also be set using `axes=dict(ytitle="my y axis")` 1920 ac : (str) 1921 axes color 1922 grid : (bool) 1923 show the background grid for the axes, can also be set using `axes=dict(xygrid=True)` 1924 ztolerance : (float) 1925 a tolerance factor to superimpose objects (along the z-axis). 1926 1927 Example: 1928 ```python 1929 from vedo.pyplot import plot 1930 import numpy as np 1931 x = np.linspace(0, 6.28, num=50) 1932 plot(np.sin(x), 'r').plot(np.cos(x), 'bo-').show().close() 1933 ``` 1934 <img src="https://user-images.githubusercontent.com/32848391/74363882-c3638300-4dcb-11ea-8a78-eb492ad9711f.png" width="600"> 1935 1936 Examples: 1937 - [plot_errbars.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/plot_errbars.py) 1938 - [plot_errband.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/plot_errband.py) 1939 - [plot_pip.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/plot_pip.py) 1940 1941  1942 1943 - [scatter1.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/scatter1.py) 1944 - [scatter2.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/scatter2.py) 1945 1946 1947 1948 ------------------------------------------------------------------------- 1949 .. note:: mode="bar" 1950 1951 Creates a `PlotBars(Figure)` object. 1952 1953 Input must be in format `[counts, labels, colors, edges]`. 1954 Either or both `edges` and `colors` are optional and can be omitted. 1955 1956 Arguments: 1957 errors : (bool) 1958 show error bars 1959 logscale : (bool) 1960 use logscale on y-axis 1961 fill : (bool) 1962 fill bars with solid color `c` 1963 gap : (float) 1964 leave a small space btw bars 1965 radius : (float) 1966 border radius of the top of the histogram bar. Default value is 0.1. 1967 texture : (str) 1968 url or path to an image to be used as texture for the bin 1969 outline : (bool) 1970 show outline of the bins 1971 xtitle : (str) 1972 title for the x-axis, can also be set using `axes=dict(xtitle="my x axis")` 1973 ytitle : (str) 1974 title for the y-axis, can also be set using `axes=dict(ytitle="my y axis")` 1975 ac : (str) 1976 axes color 1977 padding : (float, list) 1978 keep a padding space from the axes (as a fraction of the axis size). 1979 This can be a list of four numbers. 1980 aspect : (float) 1981 the desired aspect ratio of the figure. Default is 4/3. 1982 grid : (bool) 1983 show the background grid for the axes, can also be set using `axes=dict(xygrid=True)` 1984 1985 Examples: 1986 - [histo_1d_a.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_1d_a.py) 1987 - [histo_1d_b.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_1d_b.py) 1988 - [histo_1d_c.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_1d_c.py) 1989 - [histo_1d_d.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_1d_d.py) 1990 1991  1992 1993 1994 ---------------------------------------------------------------------- 1995 .. note:: 2D functions 1996 1997 If input is an external function or a formula, draw the surface 1998 representing the function `f(x,y)`. 1999 2000 Arguments: 2001 x : (float) 2002 x range of values 2003 y : (float) 2004 y range of values 2005 zlimits : (float) 2006 limit the z range of the independent variable 2007 zlevels : (int) 2008 will draw the specified number of z-levels contour lines 2009 show_nan : (bool) 2010 show where the function does not exist as red points 2011 bins : (list) 2012 number of bins in x and y 2013 2014 Examples: 2015 - [plot_fxy.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/plot_fxy.py) 2016 2017  2018 2019 2020 -------------------------------------------------------------------- 2021 .. note:: mode="complex" 2022 2023 If `mode='complex'` draw the real value of the function and color map the imaginary part. 2024 2025 Arguments: 2026 cmap : (str) 2027 diverging color map (white means `imag(z)=0`) 2028 lw : (float) 2029 line with of the binning 2030 bins : (list) 2031 binning in x and y 2032 2033 Examples: 2034 - [plot_fxy.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/plot_fxy.py) 2035 2036  2037 2038 2039 -------------------------------------------------------------------- 2040 .. note:: mode="polar" 2041 2042 If `mode='polar'` input arrays are interpreted as a list of polar angles and radii. 2043 Build a polar (radar) plot by joining the set of points in polar coordinates. 2044 2045 Arguments: 2046 title : (str) 2047 plot title 2048 tsize : (float) 2049 title size 2050 bins : (int) 2051 number of bins in phi 2052 r1 : (float) 2053 inner radius 2054 r2 : (float) 2055 outer radius 2056 lsize : (float) 2057 label size 2058 c : (color) 2059 color of the line 2060 ac : (color) 2061 color of the frame and labels 2062 alpha : (float) 2063 opacity of the frame 2064 ps : (int) 2065 point size in pixels, if ps=0 no point is drawn 2066 lw : (int) 2067 line width in pixels, if lw=0 no line is drawn 2068 deg : (bool) 2069 input array is in degrees 2070 vmax : (float) 2071 normalize radius to this maximum value 2072 fill : (bool) 2073 fill convex area with solid color 2074 splined : (bool) 2075 interpolate the set of input points 2076 show_disc : (bool) 2077 draw the outer ring axis 2078 nrays : (int) 2079 draw this number of axis rays (continuous and dashed) 2080 show_lines : (bool) 2081 draw lines to the origin 2082 show_angles : (bool) 2083 draw angle values 2084 2085 Examples: 2086 - [histo_polar.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_polar.py) 2087 2088  2089 2090 2091 -------------------------------------------------------------------- 2092 .. note:: mode="spheric" 2093 2094 If `mode='spheric'` input must be an external function rho(theta, phi). 2095 A surface is created in spherical coordinates. 2096 2097 Return an `Figure(Assembly)` of 2 objects: the unit 2098 sphere (in wireframe representation) and the surface `rho(theta, phi)`. 2099 2100 Arguments: 2101 rfunc : function 2102 handle to a user defined function `rho(theta, phi)`. 2103 normalize : (bool) 2104 scale surface to fit inside the unit sphere 2105 res : (int) 2106 grid resolution of the unit sphere 2107 scalarbar : (bool) 2108 add a 3D scalarbar to the plot for radius 2109 c : (color) 2110 color of the unit sphere 2111 alpha : (float) 2112 opacity of the unit sphere 2113 cmap : (str) 2114 color map for the surface 2115 2116 Examples: 2117 - [plot_spheric.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/plot_spheric.py) 2118 2119  2120 """ 2121 mode = kwargs.pop("mode", "") 2122 if "spher" in mode: 2123 return _plot_spheric(args[0], **kwargs) 2124 2125 if "bar" in mode: 2126 return PlotBars(args[0], **kwargs) 2127 2128 if isinstance(args[0], str) or "function" in str(type(args[0])): 2129 if "complex" in mode: 2130 return _plot_fz(args[0], **kwargs) 2131 return _plot_fxy(args[0], **kwargs) 2132 2133 # grab the matplotlib-like options 2134 optidx = None 2135 for i, a in enumerate(args): 2136 if i > 0 and isinstance(a, str): 2137 optidx = i 2138 break 2139 if optidx: 2140 opts = args[optidx].replace(" ", "") 2141 if "--" in opts: 2142 opts = opts.replace("--", "") 2143 kwargs["dashed"] = True 2144 elif "-" in opts: 2145 opts = opts.replace("-", "") 2146 else: 2147 kwargs["lw"] = 0 2148 2149 symbs = [".", "o", "O", "0", "p", "*", "h", "D", "d", "v", "^", ">", "<", "s", "x", "a"] 2150 2151 allcols = list(colors.colors.keys()) + list(colors.color_nicks.keys()) 2152 for cc in allcols: 2153 if cc == "o": 2154 continue 2155 if cc in opts: 2156 opts = opts.replace(cc, "") 2157 kwargs["lc"] = cc 2158 kwargs["mc"] = cc 2159 break 2160 2161 for ss in symbs: 2162 if ss in opts: 2163 opts = opts.replace(ss, "", 1) 2164 kwargs["marker"] = ss 2165 break 2166 2167 opts.replace(" ", "") 2168 if opts: 2169 vedo.logger.error(f"in plot(), could not understand option(s): {opts}") 2170 2171 if optidx == 1 or optidx is None: 2172 if utils.is_sequence(args[0][0]) and len(args[0][0]) > 1: 2173 # print('------------- case 1', 'plot([(x,y),..])') 2174 data = np.asarray(args[0]) # (x,y) 2175 x = np.asarray(data[:, 0]) 2176 y = np.asarray(data[:, 1]) 2177 2178 elif len(args) == 1 or optidx == 1: 2179 # print('------------- case 2', 'plot(x)') 2180 if "pandas" in str(type(args[0])): 2181 if "ytitle" not in kwargs: 2182 kwargs.update({"ytitle": args[0].name.replace("_", "_ ")}) 2183 x = np.linspace(0, len(args[0]), num=len(args[0])) 2184 y = np.asarray(args[0]).ravel() 2185 2186 elif utils.is_sequence(args[1]): 2187 # print('------------- case 3', 'plot(allx,ally)',str(type(args[0]))) 2188 if "pandas" in str(type(args[0])): 2189 if "xtitle" not in kwargs: 2190 kwargs.update({"xtitle": args[0].name.replace("_", "_ ")}) 2191 if "pandas" in str(type(args[1])): 2192 if "ytitle" not in kwargs: 2193 kwargs.update({"ytitle": args[1].name.replace("_", "_ ")}) 2194 x = np.asarray(args[0]).ravel() 2195 y = np.asarray(args[1]).ravel() 2196 2197 elif utils.is_sequence(args[0]) and utils.is_sequence(args[0][0]): 2198 # print('------------- case 4', 'plot([allx,ally])') 2199 x = np.asarray(args[0][0]).ravel() 2200 y = np.asarray(args[0][1]).ravel() 2201 2202 elif optidx == 2: 2203 # print('------------- case 5', 'plot(x,y)') 2204 x = np.asarray(args[0]).ravel() 2205 y = np.asarray(args[1]).ravel() 2206 2207 else: 2208 vedo.logger.error(f"plot(): Could not understand input arguments {args}") 2209 return None 2210 2211 if "polar" in mode: 2212 return _plot_polar(np.c_[x, y], **kwargs) 2213 2214 return PlotXY(np.c_[x, y], **kwargs)
Draw a 2D line plot, or scatter plot, of variable x vs variable y.
Input format can be either [allx], [allx, ally] or [(x1,y1), (x2,y2), ...]
Use like=... if you want to use the same format of a previously
created Figure (useful when superimposing Figures) to make sure
they are compatible and comparable. If they are not compatible
you will receive an error message.
Arguments:
- xerrors : (bool) show error bars associated to each point in x
- yerrors : (bool) show error bars associated to each point in y
- lw : (int) width of the line connecting points in pixel units. Set it to 0 to remove the line.
- lc : (str) line color
- la : (float) line "alpha", opacity of the line
- dashed : (bool) draw a dashed line instead of a continuous line
- splined : (bool) spline the line joining the point as a countinous curve
- elw : (int) width of error bar lines in units of pixels
- ec : (color) color of error bar, by default the same as marker color
- error_band : (bool)
represent errors on y as a filled error band.
Use
eckeyword to modify its color. - marker : (str, int) use a marker for the data points
- ms : (float) marker size
- mc : (color) color of the marker
- ma : (float) opacity of the marker
- xlim : (list) set limits to the range for the x variable
- ylim : (list) set limits to the range for the y variable
- aspect : (float) Desired aspect ratio. If None, it is automatically calculated to get a reasonable aspect ratio. Scaling factor is saved in Figure.yscale
- padding : (float, list) keep a padding space from the axes (as a fraction of the axis size). This can be a list of four numbers.
- title : (str) title to appear on the top of the frame, like a header.
- xtitle : (str)
title for the x-axis, can also be set using
axes=dict(xtitle="my x axis") - ytitle : (str)
title for the y-axis, can also be set using
axes=dict(ytitle="my y axis") - ac : (str) axes color
- grid : (bool)
show the background grid for the axes, can also be set using
axes=dict(xygrid=True) - ztolerance : (float) a tolerance factor to superimpose objects (along the z-axis).
Example:
from vedo.pyplot import plot import numpy as np x = np.linspace(0, 6.28, num=50) plot(np.sin(x), 'r').plot(np.cos(x), 'bo-').show().close()
Examples:
mode="bar"
Creates a PlotBars(Figure) object.
Input must be in format [counts, labels, colors, edges].
Either or both edges and colors are optional and can be omitted.
Arguments:
- errors : (bool) show error bars
- logscale : (bool) use logscale on y-axis
- fill : (bool)
fill bars with solid color
c - gap : (float) leave a small space btw bars
- radius : (float) border radius of the top of the histogram bar. Default value is 0.1.
- texture : (str) url or path to an image to be used as texture for the bin
- outline : (bool) show outline of the bins
- xtitle : (str)
title for the x-axis, can also be set using
axes=dict(xtitle="my x axis") - ytitle : (str)
title for the y-axis, can also be set using
axes=dict(ytitle="my y axis") - ac : (str) axes color
- padding : (float, list) keep a padding space from the axes (as a fraction of the axis size). This can be a list of four numbers.
- aspect : (float) the desired aspect ratio of the figure. Default is 4/3.
- grid : (bool)
show the background grid for the axes, can also be set using
axes=dict(xygrid=True)
Examples:
2D functions
If input is an external function or a formula, draw the surface
representing the function f(x,y).
Arguments:
- x : (float) x range of values
- y : (float) y range of values
- zlimits : (float) limit the z range of the independent variable
- zlevels : (int) will draw the specified number of z-levels contour lines
- show_nan : (bool) show where the function does not exist as red points
- bins : (list) number of bins in x and y
Examples:
mode="complex"
If mode='complex' draw the real value of the function and color map the imaginary part.
Arguments:
- cmap : (str)
diverging color map (white means
imag(z)=0) - lw : (float) line with of the binning
- bins : (list) binning in x and y
Examples:
mode="polar"
If mode='polar' input arrays are interpreted as a list of polar angles and radii.
Build a polar (radar) plot by joining the set of points in polar coordinates.
Arguments:
- title : (str) plot title
- tsize : (float) title size
- bins : (int) number of bins in phi
- r1 : (float) inner radius
- r2 : (float) outer radius
- lsize : (float) label size
- c : (color) color of the line
- ac : (color) color of the frame and labels
- alpha : (float) opacity of the frame
- ps : (int) point size in pixels, if ps=0 no point is drawn
- lw : (int) line width in pixels, if lw=0 no line is drawn
- deg : (bool) input array is in degrees
- vmax : (float) normalize radius to this maximum value
- fill : (bool) fill convex area with solid color
- splined : (bool) interpolate the set of input points
- show_disc : (bool) draw the outer ring axis
- nrays : (int) draw this number of axis rays (continuous and dashed)
- show_lines : (bool) draw lines to the origin
- show_angles : (bool) draw angle values
Examples:
mode="spheric"
If mode='spheric' input must be an external function rho(theta, phi).
A surface is created in spherical coordinates.
Return an Figure(Assembly) of 2 objects: the unit
sphere (in wireframe representation) and the surface rho(theta, phi).
Arguments:
- rfunc : function
handle to a user defined function
rho(theta, phi). - normalize : (bool) scale surface to fit inside the unit sphere
- res : (int) grid resolution of the unit sphere
- scalarbar : (bool) add a 3D scalarbar to the plot for radius
- c : (color) color of the unit sphere
- alpha : (float) opacity of the unit sphere
- cmap : (str) color map for the surface
Examples:
2217def histogram(*args, **kwargs): 2218 """ 2219 Histogramming for 1D and 2D data arrays. 2220 2221 This is meant as a convenience function that creates the appropriate object 2222 based on the shape of the provided input data. 2223 2224 Use keyword `like=...` if you want to use the same format of a previously 2225 created Figure (useful when superimposing Figures) to make sure 2226 they are compatible and comparable. If they are not compatible 2227 you will receive an error message. 2228 2229 ------------------------------------------------------------------------- 2230 .. note:: default mode, for 1D arrays 2231 2232 Creates a `Histogram1D(Figure)` object. 2233 2234 Arguments: 2235 weights : (list) 2236 An array of weights, of the same shape as `data`. Each value in `data` 2237 only contributes its associated weight towards the bin count (instead of 1). 2238 bins : (int) 2239 number of bins 2240 vrange : (list) 2241 restrict the range of the histogram 2242 density : (bool) 2243 normalize the area to 1 by dividing by the nr of entries and bin size 2244 logscale : (bool) 2245 use logscale on y-axis 2246 fill : (bool) 2247 fill bars with solid color `c` 2248 gap : (float) 2249 leave a small space btw bars 2250 radius : (float) 2251 border radius of the top of the histogram bar. Default value is 0.1. 2252 texture : (str) 2253 url or path to an image to be used as texture for the bin 2254 outline : (bool) 2255 show outline of the bins 2256 errors : (bool) 2257 show error bars 2258 xtitle : (str) 2259 title for the x-axis, can also be set using `axes=dict(xtitle="my x axis")` 2260 ytitle : (str) 2261 title for the y-axis, can also be set using `axes=dict(ytitle="my y axis")` 2262 padding : (float, list) 2263 keep a padding space from the axes (as a fraction of the axis size). 2264 This can be a list of four numbers. 2265 aspect : (float) 2266 the desired aspect ratio of the histogram. Default is 4/3. 2267 grid : (bool) 2268 show the background grid for the axes, can also be set using `axes=dict(xygrid=True)` 2269 ztolerance : (float) 2270 a tolerance factor to superimpose objects (along the z-axis). 2271 2272 Examples: 2273 - [histo_1d_a.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_1d_a.py) 2274 - [histo_1d_b.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_1d_b.py) 2275 - [histo_1d_c.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_1d_c.py) 2276 - [histo_1d_d.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_1d_d.py) 2277 2278  2279 2280 2281 ------------------------------------------------------------------------- 2282 .. note:: default mode, for 2D arrays 2283 2284 Input data formats `[(x1,x2,..), (y1,y2,..)] or [(x1,y1), (x2,y2),..]` 2285 are both valid. 2286 2287 Arguments: 2288 bins : (list) 2289 binning as (nx, ny) 2290 weights : (list) 2291 array of weights to assign to each entry 2292 cmap : (str, lookuptable) 2293 color map name or look up table 2294 alpha : (float) 2295 opacity of the histogram 2296 gap : (float) 2297 separation between adjacent bins as a fraction for their size. 2298 Set gap=-1 to generate a quad surface. 2299 scalarbar : (bool) 2300 add a scalarbar to right of the histogram 2301 like : (Figure) 2302 grab and use the same format of the given Figure (for superimposing) 2303 xlim : (list) 2304 [x0, x1] range of interest. If left to None will automatically 2305 choose the minimum or the maximum of the data range. 2306 Data outside the range are completely ignored. 2307 ylim : (list) 2308 [y0, y1] range of interest. If left to None will automatically 2309 choose the minimum or the maximum of the data range. 2310 Data outside the range are completely ignored. 2311 aspect : (float) 2312 the desired aspect ratio of the figure. 2313 title : (str) 2314 title of the plot to appear on top. 2315 If left blank some statistics will be shown. 2316 xtitle : (str) 2317 x axis title 2318 ytitle : (str) 2319 y axis title 2320 ztitle : (str) 2321 title for the scalar bar 2322 ac : (str) 2323 axes color, additional keyword for Axes can also be added 2324 using e.g. `axes=dict(xygrid=True)` 2325 2326 Examples: 2327 - [histo_2d_a.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_2d_a.py) 2328 - [histo_2d_b.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_2d_b.py) 2329 2330  2331 2332 2333 ------------------------------------------------------------------------- 2334 .. note:: mode="3d" 2335 2336 If `mode='3d'`, build a 2D histogram as 3D bars from a list of x and y values. 2337 2338 Arguments: 2339 xtitle : (str) 2340 x axis title 2341 bins : (int) 2342 nr of bins for the smaller range in x or y 2343 vrange : (list) 2344 range in x and y in format `[(xmin,xmax), (ymin,ymax)]` 2345 norm : (float) 2346 sets a scaling factor for the z axis (frequency axis) 2347 fill : (bool) 2348 draw solid hexagons 2349 cmap : (str) 2350 color map name for elevation 2351 gap : (float) 2352 keep a internal empty gap between bins [0,1] 2353 zscale : (float) 2354 rescale the (already normalized) zaxis for visual convenience 2355 2356 Examples: 2357 - [histo_2d_b.py](https://github.com/marcomusy/vedo/tree/master/examples/examples/pyplot/histo_2d_b.py) 2358 2359 2360 ------------------------------------------------------------------------- 2361 .. note:: mode="hexbin" 2362 2363 If `mode='hexbin'`, build a hexagonal histogram from a list of x and y values. 2364 2365 Arguments: 2366 xtitle : (str) 2367 x axis title 2368 bins : (int) 2369 nr of bins for the smaller range in x or y 2370 vrange : (list) 2371 range in x and y in format `[(xmin,xmax), (ymin,ymax)]` 2372 norm : (float) 2373 sets a scaling factor for the z axis (frequency axis) 2374 fill : (bool) 2375 draw solid hexagons 2376 cmap : (str) 2377 color map name for elevation 2378 2379 Examples: 2380 - [histo_hexagonal.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_hexagonal.py) 2381 2382  2383 2384 2385 ------------------------------------------------------------------------- 2386 .. note:: mode="polar" 2387 2388 If `mode='polar'` assume input is polar coordinate system (rho, theta): 2389 2390 Arguments: 2391 weights : (list) 2392 Array of weights, of the same shape as the input. 2393 Each value only contributes its associated weight towards the bin count (instead of 1). 2394 title : (str) 2395 histogram title 2396 tsize : (float) 2397 title size 2398 bins : (int) 2399 number of bins in phi 2400 r1 : (float) 2401 inner radius 2402 r2 : (float) 2403 outer radius 2404 phigap : (float) 2405 gap angle btw 2 radial bars, in degrees 2406 rgap : (float) 2407 gap factor along radius of numeric angle labels 2408 lpos : (float) 2409 label gap factor along radius 2410 lsize : (float) 2411 label size 2412 c : (color) 2413 color of the histogram bars, can be a list of length `bins` 2414 bc : (color) 2415 color of the frame and labels 2416 alpha : (float) 2417 opacity of the frame 2418 cmap : (str) 2419 color map name 2420 deg : (bool) 2421 input array is in degrees 2422 vmin : (float) 2423 minimum value of the radial axis 2424 vmax : (float) 2425 maximum value of the radial axis 2426 labels : (list) 2427 list of labels, must be of length `bins` 2428 show_disc : (bool) 2429 show the outer ring axis 2430 nrays : (int) 2431 draw this number of axis rays (continuous and dashed) 2432 show_lines : (bool) 2433 show lines to the origin 2434 show_angles : (bool) 2435 show angular values 2436 show_errors : (bool) 2437 show error bars 2438 2439 Examples: 2440 - [histo_polar.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_polar.py) 2441 2442  2443 2444 2445 ------------------------------------------------------------------------- 2446 .. note:: mode="spheric" 2447 2448 If `mode='spheric'`, build a histogram from list of theta and phi values. 2449 2450 Arguments: 2451 rmax : (float) 2452 maximum radial elevation of bin 2453 res : (int) 2454 sphere resolution 2455 cmap : (str) 2456 color map name 2457 lw : (int) 2458 line width of the bin edges 2459 2460 Examples: 2461 - [histo_spheric.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/histo_spheric.py) 2462 2463  2464 """ 2465 mode = kwargs.pop("mode", "") 2466 if len(args) == 2: # x, y 2467 2468 if "spher" in mode: 2469 return _histogram_spheric(args[0], args[1], **kwargs) 2470 2471 if "hex" in mode: 2472 return _histogram_hex_bin(args[0], args[1], **kwargs) 2473 2474 if "3d" in mode.lower(): 2475 return _histogram_quad_bin(args[0], args[1], **kwargs) 2476 2477 return Histogram2D(args[0], args[1], **kwargs) 2478 2479 elif len(args) == 1: 2480 2481 if isinstance(args[0], vedo.Volume): 2482 data = args[0].pointdata[0] 2483 elif isinstance(args[0], vedo.Points): 2484 pd0 = args[0].pointdata[0] 2485 if pd0: 2486 data = pd0.ravel() 2487 else: 2488 data = args[0].celldata[0].ravel() 2489 else: 2490 try: 2491 if "pandas" in str(type(args[0])): 2492 if "xtitle" not in kwargs: 2493 kwargs.update({"xtitle": args[0].name.replace("_", "_ ")}) 2494 except: 2495 pass 2496 data = np.asarray(args[0]) 2497 2498 if "spher" in mode: 2499 return _histogram_spheric(args[0][:, 0], args[0][:, 1], **kwargs) 2500 2501 if data.ndim == 1: 2502 if "polar" in mode: 2503 return _histogram_polar(data, **kwargs) 2504 return Histogram1D(data, **kwargs) 2505 2506 if "hex" in mode: 2507 return _histogram_hex_bin(args[0][:, 0], args[0][:, 1], **kwargs) 2508 2509 if "3d" in mode.lower(): 2510 return _histogram_quad_bin(args[0][:, 0], args[0][:, 1], **kwargs) 2511 2512 return Histogram2D(args[0], **kwargs) 2513 2514 vedo.logger.error(f"in histogram(): could not understand input {args[0]}") 2515 return None
Histogramming for 1D and 2D data arrays.
This is meant as a convenience function that creates the appropriate object based on the shape of the provided input data.
Use keyword like=... if you want to use the same format of a previously
created Figure (useful when superimposing Figures) to make sure
they are compatible and comparable. If they are not compatible
you will receive an error message.
default mode, for 1D arrays
Creates a Histogram1D(Figure) object.
Arguments:
- weights : (list)
An array of weights, of the same shape as
data. Each value indataonly contributes its associated weight towards the bin count (instead of 1). - bins : (int) number of bins
- vrange : (list) restrict the range of the histogram
- density : (bool) normalize the area to 1 by dividing by the nr of entries and bin size
- logscale : (bool) use logscale on y-axis
- fill : (bool)
fill bars with solid color
c - gap : (float) leave a small space btw bars
- radius : (float) border radius of the top of the histogram bar. Default value is 0.1.
- texture : (str) url or path to an image to be used as texture for the bin
- outline : (bool) show outline of the bins
- errors : (bool) show error bars
- xtitle : (str)
title for the x-axis, can also be set using
axes=dict(xtitle="my x axis") - ytitle : (str)
title for the y-axis, can also be set using
axes=dict(ytitle="my y axis") - padding : (float, list) keep a padding space from the axes (as a fraction of the axis size). This can be a list of four numbers.
- aspect : (float) the desired aspect ratio of the histogram. Default is 4/3.
- grid : (bool)
show the background grid for the axes, can also be set using
axes=dict(xygrid=True) - ztolerance : (float) a tolerance factor to superimpose objects (along the z-axis).
Examples:
default mode, for 2D arrays
Input data formats [(x1,x2,..), (y1,y2,..)] or [(x1,y1), (x2,y2),..]
are both valid.
Arguments:
- bins : (list) binning as (nx, ny)
- weights : (list) array of weights to assign to each entry
- cmap : (str, lookuptable) color map name or look up table
- alpha : (float) opacity of the histogram
- gap : (float) separation between adjacent bins as a fraction for their size. Set gap=-1 to generate a quad surface.
- scalarbar : (bool) add a scalarbar to right of the histogram
- like : (Figure) grab and use the same format of the given Figure (for superimposing)
- xlim : (list) [x0, x1] range of interest. If left to None will automatically choose the minimum or the maximum of the data range. Data outside the range are completely ignored.
- ylim : (list) [y0, y1] range of interest. If left to None will automatically choose the minimum or the maximum of the data range. Data outside the range are completely ignored.
- aspect : (float) the desired aspect ratio of the figure.
- title : (str) title of the plot to appear on top. If left blank some statistics will be shown.
- xtitle : (str) x axis title
- ytitle : (str) y axis title
- ztitle : (str) title for the scalar bar
- ac : (str)
axes color, additional keyword for Axes can also be added
using e.g.
axes=dict(xygrid=True)
Examples:
mode="3d"
If mode='3d', build a 2D histogram as 3D bars from a list of x and y values.
Arguments:
- xtitle : (str) x axis title
- bins : (int) nr of bins for the smaller range in x or y
- vrange : (list)
range in x and y in format
[(xmin,xmax), (ymin,ymax)] - norm : (float) sets a scaling factor for the z axis (frequency axis)
- fill : (bool) draw solid hexagons
- cmap : (str) color map name for elevation
- gap : (float) keep a internal empty gap between bins [0,1]
- zscale : (float) rescale the (already normalized) zaxis for visual convenience
Examples:
mode="hexbin"
If mode='hexbin', build a hexagonal histogram from a list of x and y values.
Arguments:
- xtitle : (str) x axis title
- bins : (int) nr of bins for the smaller range in x or y
- vrange : (list)
range in x and y in format
[(xmin,xmax), (ymin,ymax)] - norm : (float) sets a scaling factor for the z axis (frequency axis)
- fill : (bool) draw solid hexagons
- cmap : (str) color map name for elevation
Examples:
mode="polar"
If mode='polar' assume input is polar coordinate system (rho, theta):
Arguments:
- weights : (list) Array of weights, of the same shape as the input. Each value only contributes its associated weight towards the bin count (instead of 1).
- title : (str) histogram title
- tsize : (float) title size
- bins : (int) number of bins in phi
- r1 : (float) inner radius
- r2 : (float) outer radius
- phigap : (float) gap angle btw 2 radial bars, in degrees
- rgap : (float) gap factor along radius of numeric angle labels
- lpos : (float) label gap factor along radius
- lsize : (float) label size
- c : (color)
color of the histogram bars, can be a list of length
bins - bc : (color) color of the frame and labels
- alpha : (float) opacity of the frame
- cmap : (str) color map name
- deg : (bool) input array is in degrees
- vmin : (float) minimum value of the radial axis
- vmax : (float) maximum value of the radial axis
- labels : (list)
list of labels, must be of length
bins - show_disc : (bool) show the outer ring axis
- nrays : (int) draw this number of axis rays (continuous and dashed)
- show_lines : (bool) show lines to the origin
- show_angles : (bool) show angular values
- show_errors : (bool) show error bars
Examples:
mode="spheric"
If mode='spheric', build a histogram from list of theta and phi values.
Arguments:
- rmax : (float) maximum radial elevation of bin
- res : (int) sphere resolution
- cmap : (str) color map name
- lw : (int) line width of the bin edges
Examples:
2518def fit( 2519 points, deg=1, niter=0, nstd=3, xerrors=None, yerrors=None, vrange=None, res=250, lw=3, c="red4" 2520): 2521 """ 2522 Polynomial fitting with parameter error and error bands calculation. 2523 Errors bars in both x and y are supported. 2524 2525 Returns a `vedo.shapes.Line` object. 2526 2527 Additional information about the fitting output can be accessed with: 2528 2529 `fitd = fit(pts)` 2530 2531 - `fitd.coefficients` will contain the coefficients of the polynomial fit 2532 - `fitd.coefficient_errors`, errors on the fitting coefficients 2533 - `fitd.monte_carlo_coefficients`, fitting coefficient set from MC generation 2534 - `fitd.covariance_matrix`, covariance matrix as a numpy array 2535 - `fitd.reduced_chi2`, reduced chi-square of the fitting 2536 - `fitd.ndof`, number of degrees of freedom 2537 - `fitd.data_sigma`, mean data dispersion from the central fit assuming `Chi2=1` 2538 - `fitd.error_lines`, a `vedo.shapes.Line` object for the upper and lower error band 2539 - `fitd.error_band`, the `vedo.mesh.Mesh` object representing the error band 2540 2541 Errors on x and y can be specified. If left to `None` an estimate is made from 2542 the statistical spread of the dataset itself. Errors are always assumed gaussian. 2543 2544 Arguments: 2545 deg : (int) 2546 degree of the polynomial to be fitted 2547 niter : (int) 2548 number of monte-carlo iterations to compute error bands. 2549 If set to 0, return the simple least-squares fit with naive error estimation 2550 on coefficients only. A reasonable non-zero value to set is about 500, in 2551 this case *error_lines*, *error_band* and the other class attributes are filled 2552 nstd : (float) 2553 nr. of standard deviation to use for error calculation 2554 xerrors : (list) 2555 array of the same length of points with the errors on x 2556 yerrors : (list) 2557 array of the same length of points with the errors on y 2558 vrange : (list) 2559 specify the domain range of the fitting line 2560 (only affects visualization, but can be used to extrapolate the fit 2561 outside the data range) 2562 res : (int) 2563 resolution of the output fitted line and error lines 2564 2565 Examples: 2566 - [fit_polynomial1.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/fit_polynomial1.py) 2567 2568  2569 """ 2570 if isinstance(points, vedo.pointcloud.Points): 2571 points = points.points() 2572 points = np.asarray(points) 2573 if len(points) == 2: # assume user is passing [x,y] 2574 points = np.c_[points[0], points[1]] 2575 x = points[:, 0] 2576 y = points[:, 1] # ignore z 2577 2578 n = len(x) 2579 ndof = n - deg - 1 2580 if vrange is not None: 2581 x0, x1 = vrange 2582 else: 2583 x0, x1 = np.min(x), np.max(x) 2584 if xerrors is not None: 2585 x0 -= xerrors[0] / 2 2586 x1 += xerrors[-1] / 2 2587 2588 tol = (x1 - x0) / 10000 2589 xr = np.linspace(x0, x1, res) 2590 2591 # project x errs on y 2592 if xerrors is not None: 2593 xerrors = np.asarray(xerrors) 2594 if yerrors is not None: 2595 yerrors = np.asarray(yerrors) 2596 w = 1.0 / yerrors 2597 coeffs = np.polyfit(x, y, deg, w=w, rcond=None) 2598 else: 2599 coeffs = np.polyfit(x, y, deg, rcond=None) 2600 # update yerrors, 1 bootstrap iteration is enough 2601 p1d = np.poly1d(coeffs) 2602 der = (p1d(x + tol) - p1d(x)) / tol 2603 yerrors = np.sqrt(yerrors * yerrors + np.power(der * xerrors, 2)) 2604 2605 if yerrors is not None: 2606 yerrors = np.asarray(yerrors) 2607 w = 1.0 / yerrors 2608 coeffs, V = np.polyfit(x, y, deg, w=w, rcond=None, cov=True) 2609 else: 2610 w = 1 2611 coeffs, V = np.polyfit(x, y, deg, rcond=None, cov=True) 2612 2613 p1d = np.poly1d(coeffs) 2614 theor = p1d(xr) 2615 fitl = shapes.Line(xr, theor, lw=lw, c=c).z(tol * 2) 2616 fitl.coefficients = coeffs 2617 fitl.covariance_matrix = V 2618 residuals2_sum = np.sum(np.power(p1d(x) - y, 2)) / ndof 2619 sigma = np.sqrt(residuals2_sum) 2620 fitl.reduced_chi2 = np.sum(np.power((p1d(x) - y) * w, 2)) / ndof 2621 fitl.ndof = ndof 2622 fitl.data_sigma = sigma # worked out from data using chi2=1 hypo 2623 fitl.name = "LinearPolynomialFit" 2624 2625 if not niter: 2626 fitl.coefficient_errors = np.sqrt(np.diag(V)) 2627 return fitl ################################ 2628 2629 if yerrors is not None: 2630 sigma = yerrors 2631 else: 2632 w = None 2633 fitl.reduced_chi2 = 1 2634 2635 Theors, all_coeffs = [], [] 2636 for i in range(niter): 2637 noise = np.random.randn(n) * sigma 2638 coeffs = np.polyfit(x, y + noise, deg, w=w, rcond=None) 2639 all_coeffs.append(coeffs) 2640 P1d = np.poly1d(coeffs) 2641 Theor = P1d(xr) 2642 Theors.append(Theor) 2643 all_coeffs = np.array(all_coeffs) 2644 fitl.monte_carlo_coefficients = all_coeffs 2645 2646 stds = np.std(Theors, axis=0) 2647 fitl.coefficient_errors = np.std(all_coeffs, axis=0) 2648 2649 # check distributions on the fly 2650 # for i in range(deg+1): 2651 # histogram(all_coeffs[:,i],title='par'+str(i)).show(new=1) 2652 # histogram(all_coeffs[:,0], all_coeffs[:,1], 2653 # xtitle='param0', ytitle='param1',scalarbar=1).show(new=1) 2654 # histogram(all_coeffs[:,1], all_coeffs[:,2], 2655 # xtitle='param1', ytitle='param2').show(new=1) 2656 # histogram(all_coeffs[:,0], all_coeffs[:,2], 2657 # xtitle='param0', ytitle='param2').show(new=1) 2658 2659 error_lines = [] 2660 for i in [nstd, -nstd]: 2661 el = shapes.Line(xr, theor + stds * i, lw=1, alpha=0.2, c="k").z(tol) 2662 error_lines.append(el) 2663 el.name = "ErrorLine for sigma=" + str(i) 2664 2665 fitl.error_lines = error_lines 2666 l1 = error_lines[0].points().tolist() 2667 cband = l1 + list(reversed(error_lines[1].points().tolist())) + [l1[0]] 2668 fitl.error_band = shapes.Line(cband).triangulate().lw(0).c("k", 0.15) 2669 fitl.error_band.name = "PolynomialFitErrorBand" 2670 return fitl
Polynomial fitting with parameter error and error bands calculation. Errors bars in both x and y are supported.
Returns a vedo.shapes.Line object.
Additional information about the fitting output can be accessed with:
fitd = fit(pts)
fitd.coefficientswill contain the coefficients of the polynomial fitfitd.coefficient_errors, errors on the fitting coefficientsfitd.monte_carlo_coefficients, fitting coefficient set from MC generationfitd.covariance_matrix, covariance matrix as a numpy arrayfitd.reduced_chi2, reduced chi-square of the fittingfitd.ndof, number of degrees of freedomfitd.data_sigma, mean data dispersion from the central fit assumingChi2=1fitd.error_lines, avedo.shapes.Lineobject for the upper and lower error bandfitd.error_band, thevedo.mesh.Meshobject representing the error band
Errors on x and y can be specified. If left to None an estimate is made from
the statistical spread of the dataset itself. Errors are always assumed gaussian.
Arguments:
- deg : (int) degree of the polynomial to be fitted
- niter : (int) number of monte-carlo iterations to compute error bands. If set to 0, return the simple least-squares fit with naive error estimation on coefficients only. A reasonable non-zero value to set is about 500, in this case error_lines, error_band and the other class attributes are filled
- nstd : (float) nr. of standard deviation to use for error calculation
- xerrors : (list) array of the same length of points with the errors on x
- yerrors : (list) array of the same length of points with the errors on y
- vrange : (list) specify the domain range of the fitting line (only affects visualization, but can be used to extrapolate the fit outside the data range)
- res : (int) resolution of the output fitted line and error lines
Examples:
3394def donut( 3395 fractions, 3396 title="", 3397 tsize=0.3, 3398 r1=1.7, 3399 r2=1, 3400 phigap=0, 3401 lpos=0.8, 3402 lsize=0.15, 3403 c=None, 3404 bc="k", 3405 alpha=1, 3406 labels=(), 3407 show_disc=False, 3408): 3409 """ 3410 Donut plot or pie chart. 3411 3412 Arguments: 3413 title : (str) 3414 plot title 3415 tsize : (float) 3416 title size 3417 r1 : (float) inner radius 3418 r2 : (float) 3419 outer radius, starting from r1 3420 phigap : (float) 3421 gap angle btw 2 radial bars, in degrees 3422 lpos : (float) 3423 label gap factor along radius 3424 lsize : (float) 3425 label size 3426 c : (color) 3427 color of the plot slices 3428 bc : (color) 3429 color of the disc frame 3430 alpha : (float) 3431 opacity of the disc frame 3432 labels : (list) 3433 list of labels 3434 show_disc : (bool) 3435 show the outer ring axis 3436 3437 Examples: 3438 - [donut.py](https://github.com/marcomusy/vedo/tree/master/examples/pyplot/donut.py) 3439 3440  3441 """ 3442 fractions = np.array(fractions, dtype=float) 3443 angles = np.add.accumulate(2 * np.pi * fractions) 3444 angles[-1] = 2 * np.pi 3445 if angles[-2] > 2 * np.pi: 3446 print("Error in donut(): fractions must sum to 1.") 3447 raise RuntimeError 3448 3449 cols = [] 3450 for i, th in enumerate(np.linspace(0, 2 * np.pi, 360, endpoint=False)): 3451 for ia, a in enumerate(angles): 3452 if th < a: 3453 cols.append(c[ia]) 3454 break 3455 labs = () 3456 if labels: 3457 angles = np.concatenate([[0], angles]) 3458 labs = [""] * 360 3459 for i in range(len(labels)): 3460 a = (angles[i + 1] + angles[i]) / 2 3461 j = int(a / np.pi * 180) 3462 labs[j] = labels[i] 3463 3464 data = np.linspace(0, 2 * np.pi, 360, endpoint=False) + 0.005 3465 dn = _histogram_polar( 3466 data, 3467 title=title, 3468 bins=360, 3469 r1=r1, 3470 r2=r2, 3471 phigap=phigap, 3472 lpos=lpos, 3473 lsize=lsize, 3474 tsize=tsize, 3475 c=cols, 3476 bc=bc, 3477 alpha=alpha, 3478 vmin=0, 3479 vmax=1, 3480 labels=labs, 3481 show_disc=show_disc, 3482 show_lines=0, 3483 show_angles=0, 3484 show_errors=0, 3485 ) 3486 dn.name = "Donut" 3487 return dn
Donut plot or pie chart.
Arguments:
- title : (str) plot title
- tsize : (float) title size
- r1 : (float) inner radius
- r2 : (float) outer radius, starting from r1
- phigap : (float) gap angle btw 2 radial bars, in degrees
- lpos : (float) label gap factor along radius
- lsize : (float) label size
- c : (color) color of the plot slices
- bc : (color) color of the disc frame
- alpha : (float) opacity of the disc frame
- labels : (list) list of labels
- show_disc : (bool) show the outer ring axis
Examples:
3490def violin( 3491 values, 3492 bins=10, 3493 vlim=None, 3494 x=0, 3495 width=3, 3496 splined=True, 3497 fill=True, 3498 c="violet", 3499 alpha=1, 3500 outline=True, 3501 centerline=True, 3502 lc="darkorchid", 3503 lw=3, 3504): 3505 """ 3506 Violin style histogram. 3507 3508 Arguments: 3509 bins : (int) 3510 number of bins 3511 vlim : (list) 3512 input value limits. Crop values outside range 3513 x : (float) 3514 x-position of the violin axis 3515 width : (float) 3516 width factor of the normalized distribution 3517 splined : (bool) 3518 spline the outline 3519 fill : (bool) 3520 fill violin with solid color 3521 outline : (bool) 3522 add the distribution outline 3523 centerline : (bool) 3524 add the vertical centerline at x 3525 lc : (color) 3526 line color 3527 3528 Examples: 3529 - [histo_violin.py](https://github.com/marcomusy/vedo/tree/master/examples/examples/pyplot/histo_violin.py) 3530 3531  3532 """ 3533 fs, edges = np.histogram(values, bins=bins, range=vlim) 3534 mine, maxe = np.min(edges), np.max(edges) 3535 fs = fs.astype(float) / len(values) * width 3536 3537 rs = [] 3538 3539 if splined: 3540 lnl, lnr = [(0, edges[0], 0)], [(0, edges[0], 0)] 3541 for i in range(bins): 3542 xc = (edges[i] + edges[i + 1]) / 2 3543 yc = fs[i] 3544 lnl.append([-yc, xc, 0]) 3545 lnr.append([yc, xc, 0]) 3546 lnl.append((0, edges[-1], 0)) 3547 lnr.append((0, edges[-1], 0)) 3548 spl = shapes.KSpline(lnl).x(x) 3549 spr = shapes.KSpline(lnr).x(x) 3550 spl.color(lc).alpha(alpha).lw(lw) 3551 spr.color(lc).alpha(alpha).lw(lw) 3552 if outline: 3553 rs.append(spl) 3554 rs.append(spr) 3555 if fill: 3556 rb = shapes.Ribbon(spl, spr, c=c, alpha=alpha).lighting("off") 3557 rs.append(rb) 3558 3559 else: 3560 lns1 = [[0, mine, 0]] 3561 for i in range(bins): 3562 lns1.append([fs[i], edges[i], 0]) 3563 lns1.append([fs[i], edges[i + 1], 0]) 3564 lns1.append([0, maxe, 0]) 3565 3566 lns2 = [[0, mine, 0]] 3567 for i in range(bins): 3568 lns2.append([-fs[i], edges[i], 0]) 3569 lns2.append([-fs[i], edges[i + 1], 0]) 3570 lns2.append([0, maxe, 0]) 3571 3572 if outline: 3573 rs.append(shapes.Line(lns1, c=lc, alpha=alpha, lw=lw).x(x)) 3574 rs.append(shapes.Line(lns2, c=lc, alpha=alpha, lw=lw).x(x)) 3575 3576 if fill: 3577 for i in range(bins): 3578 p0 = (-fs[i], edges[i], 0) 3579 p1 = (fs[i], edges[i + 1], 0) 3580 r = shapes.Rectangle(p0, p1).x(p0[0] + x) 3581 r.color(c).alpha(alpha).lighting("off") 3582 rs.append(r) 3583 3584 if centerline: 3585 cl = shapes.Line([0, mine, 0.01], [0, maxe, 0.01], c=lc, alpha=alpha, lw=2).x(x) 3586 rs.append(cl) 3587 3588 asse = Assembly(rs) 3589 asse.name = "Violin" 3590 return asse
Violin style histogram.
Arguments:
- bins : (int) number of bins
- vlim : (list) input value limits. Crop values outside range
- x : (float) x-position of the violin axis
- width : (float) width factor of the normalized distribution
- splined : (bool) spline the outline
- fill : (bool) fill violin with solid color
- outline : (bool) add the distribution outline
- centerline : (bool) add the vertical centerline at x
- lc : (color) line color
Examples:
3593def whisker(data, s=0.25, c="k", lw=2, bc="blue", alpha=0.25, r=5, jitter=True, horizontal=False): 3594 """ 3595 Generate a "whisker" bar from a 1-dimensional dataset. 3596 3597 Arguments: 3598 s : (float) 3599 size of the box 3600 c : (color) 3601 color of the lines 3602 lw : (float) 3603 line width 3604 bc : (color) 3605 color of the box 3606 alpha : (float) 3607 transparency of the box 3608 r : (float) 3609 point radius in pixels (use value 0 to disable) 3610 jitter : (bool) 3611 add some randomness to points to avoid overlap 3612 horizontal : (bool) 3613 set horizontal layout 3614 3615 Examples: 3616 - [whiskers.py](https://github.com/marcomusy/vedo/tree/master/examples/examples/pyplot/whiskers.py) 3617 3618  3619 """ 3620 xvals = np.zeros_like(np.asarray(data)) 3621 if jitter: 3622 xjit = np.random.randn(len(xvals)) * s / 9 3623 xjit = np.clip(xjit, -s / 2.1, s / 2.1) 3624 xvals += xjit 3625 3626 dmean = np.mean(data) 3627 dq05 = np.quantile(data, 0.05) 3628 dq25 = np.quantile(data, 0.25) 3629 dq75 = np.quantile(data, 0.75) 3630 dq95 = np.quantile(data, 0.95) 3631 3632 pts = None 3633 if r: 3634 pts = shapes.Points([xvals, data], c=c, r=r) 3635 3636 rec = shapes.Rectangle([-s / 2, dq25], [s / 2, dq75], c=bc, alpha=alpha) 3637 rec.GetProperty().LightingOff() 3638 rl = shapes.Line([[-s / 2, dq25], [s / 2, dq25], [s / 2, dq75], [-s / 2, dq75]], closed=True) 3639 l1 = shapes.Line([0, dq05, 0], [0, dq25, 0], c=c, lw=lw) 3640 l2 = shapes.Line([0, dq75, 0], [0, dq95, 0], c=c, lw=lw) 3641 lm = shapes.Line([-s / 2, dmean], [s / 2, dmean]) 3642 lns = merge(l1, l2, lm, rl) 3643 asse = Assembly([lns, rec, pts]) 3644 if horizontal: 3645 asse.rotate_z(-90) 3646 asse.name = "Whisker" 3647 asse.info["mean"] = dmean 3648 asse.info["quantile_05"] = dq05 3649 asse.info["quantile_25"] = dq25 3650 asse.info["quantile_75"] = dq75 3651 asse.info["quantile_95"] = dq95 3652 return asse
Generate a "whisker" bar from a 1-dimensional dataset.
Arguments:
- s : (float) size of the box
- c : (color) color of the lines
- lw : (float) line width
- bc : (color) color of the box
- alpha : (float) transparency of the box
- r : (float) point radius in pixels (use value 0 to disable)
- jitter : (bool) add some randomness to points to avoid overlap
- horizontal : (bool) set horizontal layout
Examples:
3655def streamplot( 3656 X, Y, U, V, direction="both", max_propagation=None, mode=1, lw=0.001, c=None, probes=() 3657): 3658 """ 3659 Generate a streamline plot of a vectorial field (U,V) defined at positions (X,Y). 3660 Returns a `Mesh` object. 3661 3662 Arguments: 3663 direction : (str) 3664 either "forward", "backward" or "both" 3665 max_propagation : (float) 3666 maximum physical length of the streamline 3667 lw : (float) 3668 line width in absolute units 3669 mode : (int) 3670 mode of varying the line width: 3671 - 0 - do not vary line width 3672 - 1 - vary line width by first vector component 3673 - 2 - vary line width vector magnitude 3674 - 3 - vary line width by absolute value of first vector component 3675 3676 Examples: 3677 - [plot_stream.py](https://github.com/marcomusy/vedo/tree/master/examples/examples/pyplot/plot_stream.py) 3678 3679  3680 """ 3681 n = len(X) 3682 m = len(Y[0]) 3683 if n != m: 3684 print("Limitation in streamplot(): only square grids are allowed.", n, m) 3685 raise RuntimeError() 3686 3687 xmin, xmax = X[0][0], X[-1][-1] 3688 ymin, ymax = Y[0][0], Y[-1][-1] 3689 3690 field = np.sqrt(U * U + V * V) 3691 3692 vol = vedo.Volume(field, dims=(n, n, 1)) 3693 3694 uf = np.ravel(U, order="F") 3695 vf = np.ravel(V, order="F") 3696 vects = np.c_[uf, vf, np.zeros_like(uf)] 3697 vol.pointdata["StreamPlotField"] = vects 3698 3699 if len(probes) == 0: 3700 probe = shapes.Grid(pos=((n - 1) / 2, (n - 1) / 2, 0), s=(n - 1, n - 1), res=(n - 1, n - 1)) 3701 else: 3702 if isinstance(probes, vedo.Points): 3703 probes = probes.points() 3704 else: 3705 probes = np.array(probes, dtype=float) 3706 if len(probes[0]) == 2: 3707 probes = np.c_[probes[:, 0], probes[:, 1], np.zeros(len(probes))] 3708 sv = [(n - 1) / (xmax - xmin), (n - 1) / (ymax - ymin), 1] 3709 probes = probes - [xmin, ymin, 0] 3710 probes = np.multiply(probes, sv) 3711 probe = vedo.Points(probes) 3712 3713 stream = vedo.shapes.StreamLines( 3714 vol, 3715 probe, 3716 tubes={"radius": lw, "mode": mode}, 3717 lw=lw, 3718 max_propagation=max_propagation, 3719 direction=direction, 3720 ) 3721 if c is not None: 3722 stream.color(c) 3723 else: 3724 stream.add_scalarbar() 3725 stream.lighting("off") 3726 3727 stream.scale([1 / (n - 1) * (xmax - xmin), 1 / (n - 1) * (ymax - ymin), 1]) 3728 stream.shift(xmin, ymin) 3729 return stream
Generate a streamline plot of a vectorial field (U,V) defined at positions (X,Y).
Returns a Mesh object.
Arguments:
- direction : (str) either "forward", "backward" or "both"
- max_propagation : (float) maximum physical length of the streamline
- lw : (float) line width in absolute units
- mode : (int)
mode of varying the line width:
- 0 - do not vary line width
- 1 - vary line width by first vector component
- 2 - vary line width vector magnitude
- 3 - vary line width by absolute value of first vector component
Examples:
3732def matrix( 3733 M, 3734 title="Matrix", 3735 xtitle="", 3736 ytitle="", 3737 xlabels=(), 3738 ylabels=(), 3739 xrotation=0, 3740 cmap="Reds", 3741 vmin=None, 3742 vmax=None, 3743 precision=2, 3744 font="Theemim", 3745 scale=0, 3746 scalarbar=True, 3747 lc="white", 3748 lw=0, 3749 c="black", 3750 alpha=1, 3751): 3752 """ 3753 Generate a matrix, or a 2D color-coded plot with bin labels. 3754 3755 Returns an `Assembly` object. 3756 3757 Arguments: 3758 M : (list, numpy array) 3759 the input array to visualize 3760 title : (str) 3761 title of the plot 3762 xtitle : (str) 3763 title of the horizontal colmuns 3764 ytitle : (str) 3765 title of the vertical rows 3766 xlabels : (list) 3767 individual string labels for each column. Must be of length m 3768 ylabels : (list) 3769 individual string labels for each row. Must be of length n 3770 xrotation : (float) 3771 rotation of the horizontal labels 3772 cmap : (str) 3773 color map name 3774 vmin : (float) 3775 minimum value of the colormap range 3776 vmax : (float) 3777 maximum value of the colormap range 3778 precision : (int) 3779 number of digits for the matrix entries or bins 3780 font : (str) 3781 font name. Check [available fonts here](https://vedo.embl.es/fonts). 3782 3783 scale : (float) 3784 size of the numeric entries or bin values 3785 scalarbar : (bool) 3786 add a scalar bar to the right of the plot 3787 lc : (str) 3788 color of the line separating the bins 3789 lw : (float) 3790 Width of the line separating the bins 3791 c : (str) 3792 text color 3793 alpha : (float) 3794 plot transparency 3795 3796 Examples: 3797 - [np_matrix.py](https://github.com/marcomusy/vedo/tree/master/examples/examples/pyplot/np_matrix.py) 3798 3799  3800 """ 3801 M = np.asarray(M) 3802 n, m = M.shape 3803 gr = shapes.Grid(res=(m, n), s=(m / (m + n) * 2, n / (m + n) * 2), c=c, alpha=alpha) 3804 gr.wireframe(False).lc(lc).lw(lw) 3805 3806 matr = np.flip(np.flip(M), axis=1).ravel(order="C") 3807 gr.cmap(cmap, matr, on="cells", vmin=vmin, vmax=vmax) 3808 sbar = None 3809 if scalarbar: 3810 gr.add_scalarbar3d(title_font=font, label_font=font) 3811 sbar = gr.scalarbar 3812 labs = None 3813 if scale != 0: 3814 labs = gr.labels( 3815 on="cells", 3816 scale=scale / max(m, n), 3817 precision=precision, 3818 font=font, 3819 justify="center", 3820 c=c, 3821 ) 3822 labs.z(0.001) 3823 t = None 3824 if title: 3825 if title == "Matrix": 3826 title += " " + str(n) + "x" + str(m) 3827 t = shapes.Text3D(title, font=font, s=0.04, justify="bottom-center", c=c) 3828 t.shift(0, n / (m + n) * 1.05) 3829 3830 xlabs = None 3831 if len(xlabels) == m: 3832 xlabs = [] 3833 jus = "top-center" 3834 if xrotation > 44: 3835 jus = "right-center" 3836 for i in range(m): 3837 xl = shapes.Text3D(xlabels[i], font=font, s=0.02, justify=jus, c=c).rotate_z(xrotation) 3838 xl.shift((2 * i - m + 1) / (m + n), -n / (m + n) * 1.05) 3839 xlabs.append(xl) 3840 3841 ylabs = None 3842 if len(ylabels) == n: 3843 ylabels = list(reversed(ylabels)) 3844 ylabs = [] 3845 for i in range(n): 3846 yl = shapes.Text3D(ylabels[i], font=font, s=0.02, justify="right-center", c=c) 3847 yl.shift(-m / (m + n) * 1.05, (2 * i - n + 1) / (m + n)) 3848 ylabs.append(yl) 3849 3850 xt = None 3851 if xtitle: 3852 xt = shapes.Text3D(xtitle, font=font, s=0.035, justify="top-center", c=c) 3853 xt.shift(0, -n / (m + n) * 1.05) 3854 if xlabs is not None: 3855 y0, y1 = xlabs[0].ybounds() 3856 xt.shift(0, -(y1 - y0) - 0.55 / (m + n)) 3857 yt = None 3858 if ytitle: 3859 yt = shapes.Text3D(ytitle, font=font, s=0.035, justify="bottom-center", c=c).rotate_z(90) 3860 yt.shift(-m / (m + n) * 1.05, 0) 3861 if ylabs is not None: 3862 x0, x1 = ylabs[0].xbounds() 3863 yt.shift(-(x1 - x0) - 0.55 / (m + n), 0) 3864 asse = Assembly(gr, sbar, labs, t, xt, yt, xlabs, ylabs) 3865 asse.name = "Matrix" 3866 return asse
Generate a matrix, or a 2D color-coded plot with bin labels.
Returns an Assembly object.
Arguments:
- M : (list, numpy array) the input array to visualize
- title : (str) title of the plot
- xtitle : (str) title of the horizontal colmuns
- ytitle : (str) title of the vertical rows
- xlabels : (list) individual string labels for each column. Must be of length m
- ylabels : (list) individual string labels for each row. Must be of length n
- xrotation : (float) rotation of the horizontal labels
- cmap : (str) color map name
- vmin : (float) minimum value of the colormap range
- vmax : (float) maximum value of the colormap range
- precision : (int) number of digits for the matrix entries or bins
- font : (str) font name. Check available fonts here.
- scale : (float) size of the numeric entries or bin values
- scalarbar : (bool) add a scalar bar to the right of the plot
- lc : (str) color of the line separating the bins
- lw : (float) Width of the line separating the bins
- c : (str) text color
- alpha : (float) plot transparency
Examples:
4016class DirectedGraph(Assembly): 4017 """ 4018 Support for Directed Graphs. 4019 """ 4020 4021 def __init__(self, **kargs): 4022 """ 4023 A graph consists of a collection of nodes (without postional information) 4024 and a collection of edges connecting pairs of nodes. 4025 The task is to determine the node positions only based on their connections. 4026 4027 This class is derived from class `Assembly`, and it assembles 4 Mesh objects 4028 representing the graph, the node labels, edge labels and edge arrows. 4029 4030 Arguments: 4031 c : (color) 4032 Color of the Graph 4033 n : (int) 4034 number of the initial set of nodes 4035 layout : (int, str) 4036 layout in 4037 `['2d', 'fast2d', 'clustering2d', 'circular', 'circular3d', 'cone', 'force', 'tree']`. 4038 Each of these layouts has different available options. 4039 4040 --------------------------------------------------------------- 4041 .. note:: Options for layouts '2d', 'fast2d' and 'clustering2d' 4042 4043 Arguments: 4044 seed : (int) 4045 seed of the random number generator used to jitter point positions 4046 rest_distance : (float) 4047 manually set the resting distance 4048 nmax : (int) 4049 the maximum number of iterations to be used 4050 zrange : (list) 4051 expand 2d graph along z axis. 4052 4053 --------------------------------------------------------------- 4054 .. note:: Options for layouts 'circular', and 'circular3d': 4055 4056 Arguments: 4057 radius : (float) 4058 set the radius of the circles 4059 height : (float) 4060 set the vertical (local z) distance between the circles 4061 zrange : (float) 4062 expand 2d graph along z axis 4063 4064 --------------------------------------------------------------- 4065 .. note:: Options for layout 'cone' 4066 4067 Arguments: 4068 compactness : (float) 4069 ratio between the average width of a cone in the tree, 4070 and the height of the cone. 4071 compression : (bool) 4072 put children closer together, possibly allowing sub-trees to overlap. 4073 This is useful if the tree is actually the spanning tree of a graph. 4074 spacing : (float) 4075 space between layers of the tree 4076 4077 --------------------------------------------------------------- 4078 .. note:: Options for layout 'force' 4079 4080 Arguments: 4081 seed : (int) 4082 seed the random number generator used to jitter point positions 4083 bounds : (list) 4084 set the region in space in which to place the final graph 4085 nmax : (int) 4086 the maximum number of iterations to be used 4087 three_dimensional : (bool) 4088 allow optimization in the 3rd dimension too 4089 random_initial_points : (bool) 4090 use random positions within the graph bounds as initial points 4091 4092 Examples: 4093 - [lineage_graph.py](https://github.com/marcomusy/vedo/tree/master/examples/examples/pyplot/lineage_graph.py) 4094 4095  4096 4097 - [graph_network.py](https://github.com/marcomusy/vedo/tree/master/examples/examples/pyplot/graph_network.py) 4098 4099  4100 """ 4101 4102 vedo.Assembly.__init__(self) 4103 4104 self.nodes = [] 4105 self.edges = [] 4106 4107 self._node_labels = [] # holds strings 4108 self._edge_labels = [] 4109 self.edge_orientations = [] 4110 self.edge_glyph_position = 0.6 4111 4112 self.zrange = 0.0 4113 4114 self.rotX = 0 4115 self.rotY = 0 4116 self.rotZ = 0 4117 4118 self.arrow_scale = 0.15 4119 self.node_label_scale = None 4120 self.node_label_justify = "bottom-left" 4121 4122 self.edge_label_scale = None 4123 4124 self.mdg = vtk.vtkMutableDirectedGraph() 4125 4126 n = kargs.pop("n", 0) 4127 for _ in range(n): 4128 self.add_node() 4129 4130 self._c = kargs.pop("c", (0.3, 0.3, 0.3)) 4131 4132 self.gl = vtk.vtkGraphLayout() 4133 4134 self.font = kargs.pop("font", "") 4135 4136 s = kargs.pop("layout", "2d") 4137 if isinstance(s, int): 4138 ss = ["2d", "fast2d", "clustering2d", "circular", "circular3d", "cone", "force", "tree"] 4139 s = ss[s] 4140 self.layout = s 4141 4142 if "2d" in s: 4143 if "clustering" in s: 4144 self.strategy = vtk.vtkClustering2DLayoutStrategy() 4145 elif "fast" in s: 4146 self.strategy = vtk.vtkFast2DLayoutStrategy() 4147 else: 4148 self.strategy = vtk.vtkSimple2DLayoutStrategy() 4149 self.rotX = 180 4150 opt = kargs.pop("rest_distance", None) 4151 if opt is not None: 4152 self.strategy.SetRestDistance(opt) 4153 opt = kargs.pop("seed", None) 4154 if opt is not None: 4155 self.strategy.SetRandomSeed(opt) 4156 opt = kargs.pop("nmax", None) 4157 if opt is not None: 4158 self.strategy.SetMaxNumberOfIterations(opt) 4159 self.zrange = kargs.pop("zrange", 0) 4160 4161 elif "circ" in s: 4162 if "3d" in s: 4163 self.strategy = vtk.vtkSimple3DCirclesStrategy() 4164 self.strategy.SetDirection(0, 0, -1) 4165 self.strategy.SetAutoHeight(True) 4166 self.strategy.SetMethod(1) 4167 self.rotX = -90 4168 opt = kargs.pop("radius", None) # float 4169 if opt is not None: 4170 self.strategy.SetMethod(0) 4171 self.strategy.SetRadius(opt) # float 4172 opt = kargs.pop("height", None) 4173 if opt is not None: 4174 self.strategy.SetAutoHeight(False) 4175 self.strategy.SetHeight(opt) # float 4176 else: 4177 self.strategy = vtk.vtkCircularLayoutStrategy() 4178 self.zrange = kargs.pop("zrange", 0) 4179 4180 elif "cone" in s: 4181 self.strategy = vtk.vtkConeLayoutStrategy() 4182 self.rotX = 180 4183 opt = kargs.pop("compactness", None) 4184 if opt is not None: 4185 self.strategy.SetCompactness(opt) 4186 opt = kargs.pop("compression", None) 4187 if opt is not None: 4188 self.strategy.SetCompression(opt) 4189 opt = kargs.pop("spacing", None) 4190 if opt is not None: 4191 self.strategy.SetSpacing(opt) 4192 4193 elif "force" in s: 4194 self.strategy = vtk.vtkForceDirectedLayoutStrategy() 4195 opt = kargs.pop("seed", None) 4196 if opt is not None: 4197 self.strategy.SetRandomSeed(opt) 4198 opt = kargs.pop("bounds", None) 4199 if opt is not None: 4200 self.strategy.SetAutomaticBoundsComputation(False) 4201 self.strategy.SetGraphBounds(opt) # list 4202 opt = kargs.pop("nmax", None) 4203 if opt is not None: 4204 self.strategy.SetMaxNumberOfIterations(opt) # int 4205 opt = kargs.pop("three_dimensional", True) 4206 if opt is not None: 4207 self.strategy.SetThreeDimensionalLayout(opt) # bool 4208 opt = kargs.pop("random_initial_points", None) 4209 if opt is not None: 4210 self.strategy.SetRandomInitialPoints(opt) # bool 4211 4212 elif "tree" in s: 4213 self.strategy = vtk.vtkSpanTreeLayoutStrategy() 4214 self.rotX = 180 4215 4216 else: 4217 vedo.logger.error(f"Cannot understand layout {s}. Available layouts:") 4218 vedo.logger.error("[2d,fast2d,clustering2d,circular,circular3d,cone,force,tree]") 4219 raise RuntimeError() 4220 4221 self.gl.SetLayoutStrategy(self.strategy) 4222 4223 if len(kargs) > 0: 4224 vedo.logger.error(f"Cannot understand options: {kargs}") 4225 4226 def add_node(self, label="id"): 4227 """Add a new node to the `Graph`.""" 4228 v = self.mdg.AddVertex() # vtk calls it vertex.. 4229 self.nodes.append(v) 4230 if label == "id": 4231 label = int(v) 4232 self._node_labels.append(str(label)) 4233 return v 4234 4235 def add_edge(self, v1, v2, label=""): 4236 """Add a new edge between to nodes. 4237 An extra node is created automatically if needed.""" 4238 nv = len(self.nodes) 4239 if v1 >= nv: 4240 for _ in range(nv, v1 + 1): 4241 self.add_node() 4242 nv = len(self.nodes) 4243 if v2 >= nv: 4244 for _ in range(nv, v2 + 1): 4245 self.add_node() 4246 e = self.mdg.AddEdge(v1, v2) 4247 self.edges.append(e) 4248 self._edge_labels.append(str(label)) 4249 return e 4250 4251 def add_child(self, v, node_label="id", edge_label=""): 4252 """Add a new edge to a new node as its child. 4253 The extra node is created automatically if needed.""" 4254 nv = len(self.nodes) 4255 if v >= nv: 4256 for _ in range(nv, v + 1): 4257 self.add_node() 4258 child = self.mdg.AddChild(v) 4259 self.edges.append((v, child)) 4260 self.nodes.append(child) 4261 if node_label == "id": 4262 node_label = int(child) 4263 self._node_labels.append(str(node_label)) 4264 self._edge_labels.append(str(edge_label)) 4265 return child 4266 4267 def build(self): 4268 """ 4269 Build the `DirectedGraph(Assembly)`. 4270 Accessory objects are also created for labels and arrows. 4271 """ 4272 self.gl.SetZRange(self.zrange) 4273 self.gl.SetInputData(self.mdg) 4274 self.gl.Update() 4275 4276 gr2poly = vtk.vtkGraphToPolyData() 4277 gr2poly.EdgeGlyphOutputOn() 4278 gr2poly.SetEdgeGlyphPosition(self.edge_glyph_position) 4279 gr2poly.SetInputData(self.gl.GetOutput()) 4280 gr2poly.Update() 4281 4282 dgraph = Mesh(gr2poly.GetOutput(0)) 4283 # dgraph.clean() # WRONG!!! dont uncomment 4284 dgraph.flat().color(self._c).lw(2) 4285 dgraph.name = "DirectedGraph" 4286 4287 diagsz = self.diagonal_size() / 1.42 4288 if not diagsz: 4289 return None 4290 4291 dgraph.SetScale(1 / diagsz) 4292 if self.rotX: 4293 dgraph.rotate_x(self.rotX) 4294 if self.rotY: 4295 dgraph.rotate_y(self.rotY) 4296 if self.rotZ: 4297 dgraph.rotate_z(self.rotZ) 4298 4299 vecs = gr2poly.GetOutput(1).GetPointData().GetVectors() 4300 self.edge_orientations = utils.vtk2numpy(vecs) 4301 4302 # Use Glyph3D to repeat the glyph on all edges. 4303 arrows = None 4304 if self.arrow_scale: 4305 arrow_source = vtk.vtkGlyphSource2D() 4306 arrow_source.SetGlyphTypeToEdgeArrow() 4307 arrow_source.SetScale(self.arrow_scale) 4308 arrow_source.Update() 4309 arrow_glyph = vtk.vtkGlyph3D() 4310 arrow_glyph.SetInputData(0, gr2poly.GetOutput(1)) 4311 arrow_glyph.SetInputData(1, arrow_source.GetOutput()) 4312 arrow_glyph.Update() 4313 arrows = Mesh(arrow_glyph.GetOutput()) 4314 arrows.SetScale(1 / diagsz) 4315 arrows.lighting("off").color(self._c) 4316 if self.rotX: 4317 arrows.rotate_x(self.rotX) 4318 if self.rotY: 4319 arrows.rotate_y(self.rotY) 4320 if self.rotZ: 4321 arrows.rotate_z(self.rotZ) 4322 arrows.name = "DirectedGraphArrows" 4323 4324 node_labels = dgraph.labels( 4325 self._node_labels, 4326 scale=self.node_label_scale, 4327 precision=0, 4328 font=self.font, 4329 justify=self.node_label_justify, 4330 ) 4331 node_labels.color(self._c).pickable(True) 4332 node_labels.name = "DirectedGraphNodeLabels" 4333 4334 edge_labels = dgraph.labels( 4335 self._edge_labels, on="cells", scale=self.edge_label_scale, precision=0, font=self.font 4336 ) 4337 edge_labels.color(self._c).pickable(True) 4338 edge_labels.name = "DirectedGraphEdgeLabels" 4339 4340 Assembly.__init__(self, [dgraph, node_labels, edge_labels, arrows]) 4341 self.name = "DirectedGraphAssembly" 4342 return self
Support for Directed Graphs.
4021 def __init__(self, **kargs): 4022 """ 4023 A graph consists of a collection of nodes (without postional information) 4024 and a collection of edges connecting pairs of nodes. 4025 The task is to determine the node positions only based on their connections. 4026 4027 This class is derived from class `Assembly`, and it assembles 4 Mesh objects 4028 representing the graph, the node labels, edge labels and edge arrows. 4029 4030 Arguments: 4031 c : (color) 4032 Color of the Graph 4033 n : (int) 4034 number of the initial set of nodes 4035 layout : (int, str) 4036 layout in 4037 `['2d', 'fast2d', 'clustering2d', 'circular', 'circular3d', 'cone', 'force', 'tree']`. 4038 Each of these layouts has different available options. 4039 4040 --------------------------------------------------------------- 4041 .. note:: Options for layouts '2d', 'fast2d' and 'clustering2d' 4042 4043 Arguments: 4044 seed : (int) 4045 seed of the random number generator used to jitter point positions 4046 rest_distance : (float) 4047 manually set the resting distance 4048 nmax : (int) 4049 the maximum number of iterations to be used 4050 zrange : (list) 4051 expand 2d graph along z axis. 4052 4053 --------------------------------------------------------------- 4054 .. note:: Options for layouts 'circular', and 'circular3d': 4055 4056 Arguments: 4057 radius : (float) 4058 set the radius of the circles 4059 height : (float) 4060 set the vertical (local z) distance between the circles 4061 zrange : (float) 4062 expand 2d graph along z axis 4063 4064 --------------------------------------------------------------- 4065 .. note:: Options for layout 'cone' 4066 4067 Arguments: 4068 compactness : (float) 4069 ratio between the average width of a cone in the tree, 4070 and the height of the cone. 4071 compression : (bool) 4072 put children closer together, possibly allowing sub-trees to overlap. 4073 This is useful if the tree is actually the spanning tree of a graph. 4074 spacing : (float) 4075 space between layers of the tree 4076 4077 --------------------------------------------------------------- 4078 .. note:: Options for layout 'force' 4079 4080 Arguments: 4081 seed : (int) 4082 seed the random number generator used to jitter point positions 4083 bounds : (list) 4084 set the region in space in which to place the final graph 4085 nmax : (int) 4086 the maximum number of iterations to be used 4087 three_dimensional : (bool) 4088 allow optimization in the 3rd dimension too 4089 random_initial_points : (bool) 4090 use random positions within the graph bounds as initial points 4091 4092 Examples: 4093 - [lineage_graph.py](https://github.com/marcomusy/vedo/tree/master/examples/examples/pyplot/lineage_graph.py) 4094 4095  4096 4097 - [graph_network.py](https://github.com/marcomusy/vedo/tree/master/examples/examples/pyplot/graph_network.py) 4098 4099  4100 """ 4101 4102 vedo.Assembly.__init__(self) 4103 4104 self.nodes = [] 4105 self.edges = [] 4106 4107 self._node_labels = [] # holds strings 4108 self._edge_labels = [] 4109 self.edge_orientations = [] 4110 self.edge_glyph_position = 0.6 4111 4112 self.zrange = 0.0 4113 4114 self.rotX = 0 4115 self.rotY = 0 4116 self.rotZ = 0 4117 4118 self.arrow_scale = 0.15 4119 self.node_label_scale = None 4120 self.node_label_justify = "bottom-left" 4121 4122 self.edge_label_scale = None 4123 4124 self.mdg = vtk.vtkMutableDirectedGraph() 4125 4126 n = kargs.pop("n", 0) 4127 for _ in range(n): 4128 self.add_node() 4129 4130 self._c = kargs.pop("c", (0.3, 0.3, 0.3)) 4131 4132 self.gl = vtk.vtkGraphLayout() 4133 4134 self.font = kargs.pop("font", "") 4135 4136 s = kargs.pop("layout", "2d") 4137 if isinstance(s, int): 4138 ss = ["2d", "fast2d", "clustering2d", "circular", "circular3d", "cone", "force", "tree"] 4139 s = ss[s] 4140 self.layout = s 4141 4142 if "2d" in s: 4143 if "clustering" in s: 4144 self.strategy = vtk.vtkClustering2DLayoutStrategy() 4145 elif "fast" in s: 4146 self.strategy = vtk.vtkFast2DLayoutStrategy() 4147 else: 4148 self.strategy = vtk.vtkSimple2DLayoutStrategy() 4149 self.rotX = 180 4150 opt = kargs.pop("rest_distance", None) 4151 if opt is not None: 4152 self.strategy.SetRestDistance(opt) 4153 opt = kargs.pop("seed", None) 4154 if opt is not None: 4155 self.strategy.SetRandomSeed(opt) 4156 opt = kargs.pop("nmax", None) 4157 if opt is not None: 4158 self.strategy.SetMaxNumberOfIterations(opt) 4159 self.zrange = kargs.pop("zrange", 0) 4160 4161 elif "circ" in s: 4162 if "3d" in s: 4163 self.strategy = vtk.vtkSimple3DCirclesStrategy() 4164 self.strategy.SetDirection(0, 0, -1) 4165 self.strategy.SetAutoHeight(True) 4166 self.strategy.SetMethod(1) 4167 self.rotX = -90 4168 opt = kargs.pop("radius", None) # float 4169 if opt is not None: 4170 self.strategy.SetMethod(0) 4171 self.strategy.SetRadius(opt) # float 4172 opt = kargs.pop("height", None) 4173 if opt is not None: 4174 self.strategy.SetAutoHeight(False) 4175 self.strategy.SetHeight(opt) # float 4176 else: 4177 self.strategy = vtk.vtkCircularLayoutStrategy() 4178 self.zrange = kargs.pop("zrange", 0) 4179 4180 elif "cone" in s: 4181 self.strategy = vtk.vtkConeLayoutStrategy() 4182 self.rotX = 180 4183 opt = kargs.pop("compactness", None) 4184 if opt is not None: 4185 self.strategy.SetCompactness(opt) 4186 opt = kargs.pop("compression", None) 4187 if opt is not None: 4188 self.strategy.SetCompression(opt) 4189 opt = kargs.pop("spacing", None) 4190 if opt is not None: 4191 self.strategy.SetSpacing(opt) 4192 4193 elif "force" in s: 4194 self.strategy = vtk.vtkForceDirectedLayoutStrategy() 4195 opt = kargs.pop("seed", None) 4196 if opt is not None: 4197 self.strategy.SetRandomSeed(opt) 4198 opt = kargs.pop("bounds", None) 4199 if opt is not None: 4200 self.strategy.SetAutomaticBoundsComputation(False) 4201 self.strategy.SetGraphBounds(opt) # list 4202 opt = kargs.pop("nmax", None) 4203 if opt is not None: 4204 self.strategy.SetMaxNumberOfIterations(opt) # int 4205 opt = kargs.pop("three_dimensional", True) 4206 if opt is not None: 4207 self.strategy.SetThreeDimensionalLayout(opt) # bool 4208 opt = kargs.pop("random_initial_points", None) 4209 if opt is not None: 4210 self.strategy.SetRandomInitialPoints(opt) # bool 4211 4212 elif "tree" in s: 4213 self.strategy = vtk.vtkSpanTreeLayoutStrategy() 4214 self.rotX = 180 4215 4216 else: 4217 vedo.logger.error(f"Cannot understand layout {s}. Available layouts:") 4218 vedo.logger.error("[2d,fast2d,clustering2d,circular,circular3d,cone,force,tree]") 4219 raise RuntimeError() 4220 4221 self.gl.SetLayoutStrategy(self.strategy) 4222 4223 if len(kargs) > 0: 4224 vedo.logger.error(f"Cannot understand options: {kargs}")
A graph consists of a collection of nodes (without postional information) and a collection of edges connecting pairs of nodes. The task is to determine the node positions only based on their connections.
This class is derived from class Assembly, and it assembles 4 Mesh objects
representing the graph, the node labels, edge labels and edge arrows.
Arguments:
- c : (color) Color of the Graph
- n : (int) number of the initial set of nodes
- layout : (int, str)
layout in
['2d', 'fast2d', 'clustering2d', 'circular', 'circular3d', 'cone', 'force', 'tree']. Each of these layouts has different available options.
Options for layouts '2d', 'fast2d' and 'clustering2d'
Arguments:
- seed : (int) seed of the random number generator used to jitter point positions
- rest_distance : (float) manually set the resting distance
- nmax : (int) the maximum number of iterations to be used
- zrange : (list) expand 2d graph along z axis.
Options for layouts 'circular', and 'circular3d':
Arguments:
- radius : (float) set the radius of the circles
- height : (float) set the vertical (local z) distance between the circles
- zrange : (float) expand 2d graph along z axis
Options for layout 'cone'
Arguments:
- compactness : (float) ratio between the average width of a cone in the tree, and the height of the cone.
- compression : (bool) put children closer together, possibly allowing sub-trees to overlap. This is useful if the tree is actually the spanning tree of a graph.
- spacing : (float) space between layers of the tree
Options for layout 'force'
Arguments:
- seed : (int) seed the random number generator used to jitter point positions
- bounds : (list) set the region in space in which to place the final graph
- nmax : (int) the maximum number of iterations to be used
- three_dimensional : (bool) allow optimization in the 3rd dimension too
- random_initial_points : (bool) use random positions within the graph bounds as initial points
Examples:
4226 def add_node(self, label="id"): 4227 """Add a new node to the `Graph`.""" 4228 v = self.mdg.AddVertex() # vtk calls it vertex.. 4229 self.nodes.append(v) 4230 if label == "id": 4231 label = int(v) 4232 self._node_labels.append(str(label)) 4233 return v
Add a new node to the Graph.
4235 def add_edge(self, v1, v2, label=""): 4236 """Add a new edge between to nodes. 4237 An extra node is created automatically if needed.""" 4238 nv = len(self.nodes) 4239 if v1 >= nv: 4240 for _ in range(nv, v1 + 1): 4241 self.add_node() 4242 nv = len(self.nodes) 4243 if v2 >= nv: 4244 for _ in range(nv, v2 + 1): 4245 self.add_node() 4246 e = self.mdg.AddEdge(v1, v2) 4247 self.edges.append(e) 4248 self._edge_labels.append(str(label)) 4249 return e
Add a new edge between to nodes. An extra node is created automatically if needed.
4251 def add_child(self, v, node_label="id", edge_label=""): 4252 """Add a new edge to a new node as its child. 4253 The extra node is created automatically if needed.""" 4254 nv = len(self.nodes) 4255 if v >= nv: 4256 for _ in range(nv, v + 1): 4257 self.add_node() 4258 child = self.mdg.AddChild(v) 4259 self.edges.append((v, child)) 4260 self.nodes.append(child) 4261 if node_label == "id": 4262 node_label = int(child) 4263 self._node_labels.append(str(node_label)) 4264 self._edge_labels.append(str(edge_label)) 4265 return child
Add a new edge to a new node as its child. The extra node is created automatically if needed.
4267 def build(self): 4268 """ 4269 Build the `DirectedGraph(Assembly)`. 4270 Accessory objects are also created for labels and arrows. 4271 """ 4272 self.gl.SetZRange(self.zrange) 4273 self.gl.SetInputData(self.mdg) 4274 self.gl.Update() 4275 4276 gr2poly = vtk.vtkGraphToPolyData() 4277 gr2poly.EdgeGlyphOutputOn() 4278 gr2poly.SetEdgeGlyphPosition(self.edge_glyph_position) 4279 gr2poly.SetInputData(self.gl.GetOutput()) 4280 gr2poly.Update() 4281 4282 dgraph = Mesh(gr2poly.GetOutput(0)) 4283 # dgraph.clean() # WRONG!!! dont uncomment 4284 dgraph.flat().color(self._c).lw(2) 4285 dgraph.name = "DirectedGraph" 4286 4287 diagsz = self.diagonal_size() / 1.42 4288 if not diagsz: 4289 return None 4290 4291 dgraph.SetScale(1 / diagsz) 4292 if self.rotX: 4293 dgraph.rotate_x(self.rotX) 4294 if self.rotY: 4295 dgraph.rotate_y(self.rotY) 4296 if self.rotZ: 4297 dgraph.rotate_z(self.rotZ) 4298 4299 vecs = gr2poly.GetOutput(1).GetPointData().GetVectors() 4300 self.edge_orientations = utils.vtk2numpy(vecs) 4301 4302 # Use Glyph3D to repeat the glyph on all edges. 4303 arrows = None 4304 if self.arrow_scale: 4305 arrow_source = vtk.vtkGlyphSource2D() 4306 arrow_source.SetGlyphTypeToEdgeArrow() 4307 arrow_source.SetScale(self.arrow_scale) 4308 arrow_source.Update() 4309 arrow_glyph = vtk.vtkGlyph3D() 4310 arrow_glyph.SetInputData(0, gr2poly.GetOutput(1)) 4311 arrow_glyph.SetInputData(1, arrow_source.GetOutput()) 4312 arrow_glyph.Update() 4313 arrows = Mesh(arrow_glyph.GetOutput()) 4314 arrows.SetScale(1 / diagsz) 4315 arrows.lighting("off").color(self._c) 4316 if self.rotX: 4317 arrows.rotate_x(self.rotX) 4318 if self.rotY: 4319 arrows.rotate_y(self.rotY) 4320 if self.rotZ: 4321 arrows.rotate_z(self.rotZ) 4322 arrows.name = "DirectedGraphArrows" 4323 4324 node_labels = dgraph.labels( 4325 self._node_labels, 4326 scale=self.node_label_scale, 4327 precision=0, 4328 font=self.font, 4329 justify=self.node_label_justify, 4330 ) 4331 node_labels.color(self._c).pickable(True) 4332 node_labels.name = "DirectedGraphNodeLabels" 4333 4334 edge_labels = dgraph.labels( 4335 self._edge_labels, on="cells", scale=self.edge_label_scale, precision=0, font=self.font 4336 ) 4337 edge_labels.color(self._c).pickable(True) 4338 edge_labels.name = "DirectedGraphEdgeLabels" 4339 4340 Assembly.__init__(self, [dgraph, node_labels, edge_labels, arrows]) 4341 self.name = "DirectedGraphAssembly" 4342 return self
Build the DirectedGraph(Assembly).
Accessory objects are also created for labels and arrows.





















