vedo.utils
Utilities submodule.
1#!/usr/bin/env python3 2# -*- coding: utf-8 -*- 3import os 4import time 5from typing import Union, Tuple, MutableSequence, List 6import numpy as np 7 8from vtkmodules.util.numpy_support import numpy_to_vtk, vtk_to_numpy 9from vtkmodules.util.numpy_support import numpy_to_vtkIdTypeArray 10import vedo.vtkclasses as vtki 11 12import vedo 13 14 15__docformat__ = "google" 16 17__doc__ = "Utilities submodule." 18 19__all__ = [ 20 "OperationNode", 21 "ProgressBar", 22 "progressbar", 23 "Minimizer", 24 "compute_hessian", 25 "geometry", 26 "is_sequence", 27 "lin_interpolate", 28 "vector", 29 "mag", 30 "mag2", 31 "versor", 32 "precision", 33 "round_to_digit", 34 "point_in_triangle", 35 "point_line_distance", 36 "closest", 37 "grep", 38 "make_bands", 39 "pack_spheres", 40 "humansort", 41 "print_histogram", 42 "print_inheritance_tree", 43 "camera_from_quaternion", 44 "camera_from_neuroglancer", 45 "camera_from_dict", 46 "camera_to_dict", 47 "oriented_camera", 48 "vedo2trimesh", 49 "trimesh2vedo", 50 "vedo2meshlab", 51 "meshlab2vedo", 52 "vedo2open3d", 53 "open3d2vedo", 54 "vtk2numpy", 55 "numpy2vtk", 56 "get_uv", 57 "andrews_curves", 58] 59 60########################################################################### 61class OperationNode: 62 """ 63 Keep track of the operations which led to a final state. 64 """ 65 # https://www.graphviz.org/doc/info/shapes.html#html 66 # Mesh #e9c46a 67 # Follower #d9ed92 68 # Volume, UnstructuredGrid #4cc9f0 69 # TetMesh #9e2a2b 70 # File #8a817c 71 # Image #f28482 72 # Assembly #f08080 73 74 def __init__( 75 self, operation, parents=(), comment="", shape="none", c="#e9c46a", style="filled" 76 ) -> None: 77 """ 78 Keep track of the operations which led to a final object. 79 This allows to show the `pipeline` tree for any `vedo` object with e.g.: 80 81 ```python 82 from vedo import * 83 sp = Sphere() 84 sp.clean().subdivide() 85 sp.pipeline.show() 86 ``` 87 88 Arguments: 89 operation : (str, class) 90 descriptor label, if a class is passed then grab its name 91 parents : (list) 92 list of the parent classes the object comes from 93 comment : (str) 94 a second-line text description 95 shape : (str) 96 shape of the frame, check out [this link.](https://graphviz.org/doc/info/shapes.html) 97 c : (hex) 98 hex color 99 style : (str) 100 comma-separated list of styles 101 102 Example: 103 ```python 104 from vedo.utils import OperationNode 105 106 op_node1 = OperationNode("Operation1", c="lightblue") 107 op_node2 = OperationNode("Operation2") 108 op_node3 = OperationNode("Operation3", shape='diamond') 109 op_node4 = OperationNode("Operation4") 110 op_node5 = OperationNode("Operation5") 111 op_node6 = OperationNode("Result", c="lightgreen") 112 113 op_node3.add_parent(op_node1) 114 op_node4.add_parent(op_node1) 115 op_node3.add_parent(op_node2) 116 op_node5.add_parent(op_node2) 117 op_node6.add_parent(op_node3) 118 op_node6.add_parent(op_node5) 119 op_node6.add_parent(op_node1) 120 121 op_node6.show(orientation="TB") 122 ``` 123 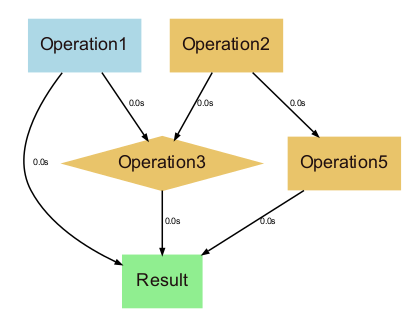 124 """ 125 if not vedo.settings.enable_pipeline: 126 return 127 128 if isinstance(operation, str): 129 self.operation = operation 130 else: 131 self.operation = operation.__class__.__name__ 132 self.operation_plain = str(self.operation) 133 134 pp = [] # filter out invalid stuff 135 for p in parents: 136 if hasattr(p, "pipeline"): 137 pp.append(p.pipeline) 138 self.parents = pp 139 140 if comment: 141 self.operation = f"<{self.operation}<BR/><SUB><I>{comment}</I></SUB>>" 142 143 self.dot = None 144 self.time = time.time() 145 self.shape = shape 146 self.style = style 147 self.color = c 148 self.counts = 0 149 150 def add_parent(self, parent) -> None: 151 """Add a parent to the list.""" 152 self.parents.append(parent) 153 154 def _build_tree(self, dot): 155 dot.node( 156 str(id(self)), 157 label=self.operation, 158 shape=self.shape, 159 color=self.color, 160 style=self.style, 161 ) 162 for parent in self.parents: 163 if parent: 164 t = f"{self.time - parent.time: .1f}s" 165 dot.edge(str(id(parent)), str(id(self)), label=t) 166 parent._build_tree(dot) 167 168 def __repr__(self): 169 try: 170 from treelib import Tree 171 except ImportError: 172 vedo.logger.error( 173 "To use this functionality please install treelib:" 174 "\n pip install treelib" 175 ) 176 return "" 177 178 def _build_tree(parent): 179 for par in parent.parents: 180 if par: 181 op = par.operation_plain 182 tree.create_node( 183 op, op + str(par.time), parent=parent.operation_plain + str(parent.time) 184 ) 185 _build_tree(par) 186 try: 187 tree = Tree() 188 tree.create_node(self.operation_plain, self.operation_plain + str(self.time)) 189 _build_tree(self) 190 out = tree.show(stdout=False) 191 except: 192 out = f"Sorry treelib failed to build the tree for '{self.operation_plain}()'." 193 return out 194 195 def print(self) -> None: 196 """Print the tree of operations.""" 197 print(self.__str__()) 198 199 def show(self, orientation="LR", popup=True) -> None: 200 """Show the graphviz output for the pipeline of this object""" 201 if not vedo.settings.enable_pipeline: 202 return 203 204 try: 205 from graphviz import Digraph 206 except ImportError: 207 vedo.logger.error("please install graphviz with command\n pip install graphviz") 208 vedo.logger.error(" sudo apt-get install graphviz -y") 209 return 210 211 # visualize the entire tree 212 dot = Digraph( 213 node_attr={"fontcolor": "#201010", "fontname": "Helvetica", "fontsize": "12"}, 214 edge_attr={"fontname": "Helvetica", "fontsize": "6", "arrowsize": "0.4"}, 215 ) 216 dot.attr(rankdir=orientation) 217 218 self.counts = 0 219 self._build_tree(dot) 220 self.dot = dot 221 222 home_dir = os.path.expanduser("~") 223 gpath = os.path.join( 224 home_dir, vedo.settings.cache_directory, "vedo", "pipeline_graphviz") 225 226 dot.render(gpath, view=popup) 227 228 229########################################################################### 230class ProgressBar: 231 """ 232 Class to print a progress bar. 233 """ 234 235 def __init__( 236 self, 237 start, 238 stop, 239 step=1, 240 c=None, 241 bold=True, 242 italic=False, 243 title="", 244 eta=True, 245 delay=-1, 246 width=25, 247 char="\U00002501", 248 char_back="\U00002500", 249 ) -> None: 250 """ 251 Class to print a progress bar with optional text message. 252 253 Check out also function `progressbar()`. 254 255 Arguments: 256 start : (int) 257 starting value 258 stop : (int) 259 stopping value 260 step : (int) 261 step value 262 c : (str) 263 color in hex format 264 title : (str) 265 title text 266 eta : (bool) 267 estimate time of arrival 268 delay : (float) 269 minimum time before printing anything, 270 if negative use the default value 271 as set in `vedo.settings.progressbar_delay` 272 width : (int) 273 width of the progress bar 274 char : (str) 275 character to use for the progress bar 276 char_back : (str) 277 character to use for the background of the progress bar 278 279 Example: 280 ```python 281 import time 282 from vedo import ProgressBar 283 pb = ProgressBar(0,40, c='r') 284 for i in pb.range(): 285 time.sleep(0.1) 286 pb.print() 287 ``` 288  289 """ 290 self.char = char 291 self.char_back = char_back 292 293 self.title = title + " " 294 if title: 295 self.title = " " + self.title 296 297 if delay < 0: 298 delay = vedo.settings.progressbar_delay 299 300 self.start = start 301 self.stop = stop 302 self.step = step 303 304 self.color = c 305 self.bold = bold 306 self.italic = italic 307 self.width = width 308 self.pbar = "" 309 self.percent = 0.0 310 self.percent_int = 0 311 self.eta = eta 312 self.delay = delay 313 314 self.t0 = time.time() 315 self._remaining = 1e10 316 317 self._update(0) 318 319 self._counts = 0 320 self._oldbar = "" 321 self._lentxt = 0 322 self._range = np.arange(start, stop, step) 323 324 def print(self, txt="", c=None) -> None: 325 """Print the progress bar with an optional message.""" 326 if not c: 327 c = self.color 328 329 self._update(self._counts + self.step) 330 331 if self.delay: 332 if time.time() - self.t0 < self.delay: 333 return 334 335 if self.pbar != self._oldbar: 336 self._oldbar = self.pbar 337 338 if self.eta and self._counts > 1: 339 340 tdenom = time.time() - self.t0 341 if tdenom: 342 vel = self._counts / tdenom 343 self._remaining = (self.stop - self._counts) / vel 344 else: 345 vel = 1 346 self._remaining = 0.0 347 348 if self._remaining > 60: 349 _mins = int(self._remaining / 60) 350 _secs = self._remaining - 60 * _mins 351 mins = f"{_mins}m" 352 secs = f"{int(_secs + 0.5)}s " 353 else: 354 mins = "" 355 secs = f"{int(self._remaining + 0.5)}s " 356 357 vel = round(vel, 1) 358 eta = f"eta: {mins}{secs}({vel} it/s) " 359 if self._remaining < 0.5: 360 dt = time.time() - self.t0 361 if dt > 60: 362 _mins = int(dt / 60) 363 _secs = dt - 60 * _mins 364 mins = f"{_mins}m" 365 secs = f"{int(_secs + 0.5)}s " 366 else: 367 mins = "" 368 secs = f"{int(dt + 0.5)}s " 369 eta = f"elapsed: {mins}{secs}({vel} it/s) " 370 txt = "" 371 else: 372 eta = "" 373 374 eraser = " " * self._lentxt + "\b" * self._lentxt 375 376 s = f"{self.pbar} {eraser}{eta}{txt}\r" 377 vedo.printc(s, c=c, bold=self.bold, italic=self.italic, end="") 378 if self.percent > 99.999: 379 print("") 380 381 self._lentxt = len(txt) 382 383 def range(self) -> np.ndarray: 384 """Return the range iterator.""" 385 return self._range 386 387 def _update(self, counts): 388 if counts < self.start: 389 counts = self.start 390 elif counts > self.stop: 391 counts = self.stop 392 self._counts = counts 393 394 self.percent = (self._counts - self.start) * 100.0 395 396 delta = self.stop - self.start 397 if delta: 398 self.percent /= delta 399 else: 400 self.percent = 0.0 401 402 self.percent_int = int(round(self.percent)) 403 af = self.width - 2 404 nh = int(round(self.percent_int / 100 * af)) 405 pbar_background = "\x1b[2m" + self.char_back * (af - nh) 406 self.pbar = f"{self.title}{self.char * (nh-1)}{pbar_background}" 407 if self.percent < 100.0: 408 ps = f" {self.percent_int}%" 409 else: 410 ps = "" 411 self.pbar += ps 412 413 414##################################### 415def progressbar( 416 iterable, 417 c=None, bold=True, italic=False, title="", 418 eta=True, width=25, delay=-1, 419 ): 420 """ 421 Function to print a progress bar with optional text message. 422 423 Use delay to set a minimum time before printing anything. 424 If delay is negative, then use the default value 425 as set in `vedo.settings.progressbar_delay`. 426 427 Arguments: 428 start : (int) 429 starting value 430 stop : (int) 431 stopping value 432 step : (int) 433 step value 434 c : (str) 435 color in hex format 436 title : (str) 437 title text 438 eta : (bool) 439 estimate time of arrival 440 delay : (float) 441 minimum time before printing anything, 442 if negative use the default value 443 set in `vedo.settings.progressbar_delay` 444 width : (int) 445 width of the progress bar 446 char : (str) 447 character to use for the progress bar 448 char_back : (str) 449 character to use for the background of the progress bar 450 451 Example: 452 ```python 453 import time 454 for i in progressbar(range(100), c='r'): 455 time.sleep(0.1) 456 ``` 457  458 """ 459 try: 460 if is_number(iterable): 461 total = int(iterable) 462 iterable = range(total) 463 else: 464 total = len(iterable) 465 except TypeError: 466 iterable = list(iterable) 467 total = len(iterable) 468 469 pb = ProgressBar( 470 0, total, c=c, bold=bold, italic=italic, title=title, 471 eta=eta, delay=delay, width=width, 472 ) 473 for item in iterable: 474 pb.print() 475 yield item 476 477 478########################################################### 479class Minimizer: 480 """ 481 A function minimizer that uses the Nelder-Mead method. 482 483 The algorithm constructs an n-dimensional simplex in parameter 484 space (i.e. a tetrahedron if the number or parameters is 3) 485 and moves the vertices around parameter space until 486 a local minimum is found. The amoeba method is robust, 487 reasonably efficient, but is not guaranteed to find 488 the global minimum if several local minima exist. 489 490 Arguments: 491 function : (callable) 492 the function to minimize 493 max_iterations : (int) 494 the maximum number of iterations 495 contraction_ratio : (float) 496 The contraction ratio. 497 The default value of 0.5 gives fast convergence, 498 but larger values such as 0.6 or 0.7 provide greater stability. 499 expansion_ratio : (float) 500 The expansion ratio. 501 The default value is 2.0, which provides rapid expansion. 502 Values between 1.1 and 2.0 are valid. 503 tol : (float) 504 the tolerance for convergence 505 506 Example: 507 - [nelder-mead.py](https://github.com/marcomusy/vedo/blob/master/examples/others/nelder-mead.py) 508 """ 509 def __init__( 510 self, 511 function=None, 512 max_iterations=10000, 513 contraction_ratio=0.5, 514 expansion_ratio=2.0, 515 tol=1e-5, 516 ) -> None: 517 self.function = function 518 self.tolerance = tol 519 self.contraction_ratio = contraction_ratio 520 self.expansion_ratio = expansion_ratio 521 self.max_iterations = max_iterations 522 self.minimizer = vtki.new("AmoebaMinimizer") 523 self.minimizer.SetFunction(self._vtkfunc) 524 self.results = {} 525 self.parameters_path = [] 526 self.function_path = [] 527 528 def _vtkfunc(self): 529 n = self.minimizer.GetNumberOfParameters() 530 ain = [self.minimizer.GetParameterValue(i) for i in range(n)] 531 r = self.function(ain) 532 self.minimizer.SetFunctionValue(r) 533 self.parameters_path.append(ain) 534 self.function_path.append(r) 535 return r 536 537 def eval(self, parameters=()) -> float: 538 """ 539 Evaluate the function at the current or given parameters. 540 """ 541 if len(parameters) == 0: 542 return self.minimizer.EvaluateFunction() 543 self.set_parameters(parameters) 544 return self.function(parameters) 545 546 def set_parameter(self, name, value, scale=1.0) -> None: 547 """ 548 Set the parameter value. 549 The initial amount by which the parameter 550 will be modified during the search for the minimum. 551 """ 552 self.minimizer.SetParameterValue(name, value) 553 self.minimizer.SetParameterScale(name, scale) 554 555 def set_parameters(self, parameters) -> None: 556 """ 557 Set the parameters names and values from a dictionary. 558 """ 559 for name, value in parameters.items(): 560 if len(value) == 2: 561 self.set_parameter(name, value[0], value[1]) 562 else: 563 self.set_parameter(name, value) 564 565 def minimize(self) -> dict: 566 """ 567 Minimize the input function. 568 569 A dictionary with the minimization results 570 including the initial parameters, final parameters, 571 minimum value, number of iterations, maximum iterations, 572 tolerance, convergence flag, parameters path, 573 function path, Hessian matrix, and parameter errors. 574 575 Arguments: 576 init_parameters : (dict) 577 the initial parameters 578 parameters : (dict) 579 the final parameters 580 min_value : (float) 581 the minimum value 582 iterations : (int) 583 the number of iterations 584 max_iterations : (int) 585 the maximum number of iterations 586 tolerance : (float) 587 the tolerance for convergence 588 convergence_flag : (int) 589 zero if the tolerance stopping criterion has been met. 590 parameters_path : (np.array) 591 the path of the minimization algorithm in parameter space 592 function_path : (np.array) 593 the path of the minimization algorithm in function space 594 hessian : (np.array) 595 the Hessian matrix of the function at the minimum 596 parameter_errors : (np.array) 597 the errors on the parameters 598 """ 599 n = self.minimizer.GetNumberOfParameters() 600 out = [( 601 self.minimizer.GetParameterName(i), 602 (self.minimizer.GetParameterValue(i), 603 self.minimizer.GetParameterScale(i)) 604 ) for i in range(n)] 605 self.results["init_parameters"] = dict(out) 606 607 self.minimizer.SetTolerance(self.tolerance) 608 self.minimizer.SetContractionRatio(self.contraction_ratio) 609 self.minimizer.SetExpansionRatio(self.expansion_ratio) 610 self.minimizer.SetMaxIterations(self.max_iterations) 611 612 self.minimizer.Minimize() 613 self.results["convergence_flag"] = not bool(self.minimizer.Iterate()) 614 615 out = [( 616 self.minimizer.GetParameterName(i), 617 self.minimizer.GetParameterValue(i), 618 ) for i in range(n)] 619 620 self.results["parameters"] = dict(out) 621 self.results["min_value"] = self.minimizer.GetFunctionValue() 622 self.results["iterations"] = self.minimizer.GetIterations() 623 self.results["max_iterations"] = self.minimizer.GetMaxIterations() 624 self.results["tolerance"] = self.minimizer.GetTolerance() 625 self.results["expansion_ratio"] = self.expansion_ratio 626 self.results["contraction_ratio"] = self.contraction_ratio 627 self.results["parameters_path"] = np.array(self.parameters_path) 628 self.results["function_path"] = np.array(self.function_path) 629 self.results["hessian"] = np.zeros((n,n)) 630 self.results["parameter_errors"] = np.zeros(n) 631 return self.results 632 633 @property 634 def x(self): 635 """Return the final parameters.""" 636 return self.results["parameters"] 637 638 @property 639 def fun(self): 640 """Return the final score value.""" 641 return self.results["min_value"] 642 643 def compute_hessian(self, epsilon=0) -> np.ndarray: 644 """ 645 Compute the Hessian matrix of `function` at the 646 minimum numerically. 647 648 Arguments: 649 epsilon : (float) 650 Step size used for numerical approximation. 651 652 Returns: 653 Hessian matrix of `function` at minimum. 654 """ 655 if not epsilon: 656 epsilon = self.tolerance * 10 657 n = self.minimizer.GetNumberOfParameters() 658 x0 = [self.minimizer.GetParameterValue(i) for i in range(n)] 659 hessian = compute_hessian(self.function, x0, epsilon=epsilon) 660 661 self.results["hessian"] = hessian 662 try: 663 ihess = np.linalg.inv(hessian) 664 cov = ihess / 2 665 self.results["parameter_errors"] = np.sqrt(np.diag(cov)) 666 print(self.results["parameter_errors"]) 667 except: 668 vedo.logger.warning("Cannot compute hessian for parameter errors") 669 self.results["parameter_errors"] = np.zeros(n) 670 return hessian 671 672 def __str__(self) -> str: 673 out = vedo.printc( 674 f"vedo.utils.Minimizer at ({hex(id(self))})".ljust(75), 675 bold=True, invert=True, return_string=True, 676 ) 677 out += "Function name".ljust(20) + self.function.__name__ + "()\n" 678 out += "-------- parameters initial value -----------\n" 679 out += "Name".ljust(20) + "Value".ljust(20) + "Scale\n" 680 for name, value in self.results["init_parameters"].items(): 681 out += name.ljust(20) + str(value[0]).ljust(20) + str(value[1]) + "\n" 682 out += "-------- parameters final value --------------\n" 683 for name, value in self.results["parameters"].items(): 684 out += name.ljust(20) + f"{value:.6f}" 685 ierr = list(self.results["parameters"]).index(name) 686 err = self.results["parameter_errors"][ierr] 687 if err: 688 out += f" ± {err:.4f}" 689 out += "\n" 690 out += "Value at minimum".ljust(20)+ f'{self.results["min_value"]}\n' 691 out += "Iterations".ljust(20) + f'{self.results["iterations"]}\n' 692 out += "Max iterations".ljust(20) + f'{self.results["max_iterations"]}\n' 693 out += "Convergence flag".ljust(20)+ f'{self.results["convergence_flag"]}\n' 694 out += "Tolerance".ljust(20) + f'{self.results["tolerance"]}\n' 695 try: 696 arr = np.array2string( 697 self.compute_hessian(), 698 separator=', ', precision=6, suppress_small=True, 699 ) 700 out += "Hessian Matrix:\n" + arr 701 except: 702 out += "Hessian Matrix: (not available)" 703 return out 704 705 706def compute_hessian(func, params, bounds=None, epsilon=1e-5, verbose=True) -> np.ndarray: 707 """ 708 Compute the Hessian matrix of a scalar function `func` at `params`, 709 accounting for parameter boundaries. 710 711 Arguments: 712 func : (callable) 713 Function returning a scalar. Takes `params` as input. 714 params : (np.ndarray) 715 Parameter vector at which to compute the Hessian. 716 bounds : (list of tuples) 717 Optional bounds for parameters, e.g., [(lb1, ub1), ...]. 718 epsilon : (float) 719 Base step size for finite differences. 720 verbose : (bool) 721 Whether to print progress. 722 723 Returns: 724 Hessian matrix of shape (n_params, n_params). 725 726 Example: 727 ```python 728 from vedo import * 729 import numpy as np 730 731 # 1. Define the objective function to minimize 732 def cost_function(params): 733 a, b = params[0], params[1] 734 score = (a-5)**2 + (b-2)**2 #+a*b 735 #print(a, b, score) 736 return score 737 738 # 2. Fit parameters (example result) 739 mle_params = np.array([4.97, 1.95]) # Assume obtained via optimization 740 741 # 3. Define bounds for parameters 742 bounds = [(-np.inf, np.inf), (1e-6, 2.1)] 743 744 # 4. Compute Hessian 745 hessian = compute_hessian(cost_function, mle_params, bounds=bounds) 746 cov_matrix = np.linalg.inv(hessian) / 2 747 print("Covariance Matrix:", cov_matrix) 748 std = np.sqrt(np.diag(cov_matrix)) 749 print("Standard deviations:", std) 750 ``` 751 """ 752 n = len(params) 753 hessian = np.zeros((n, n)) 754 f_0 = func(params) # Central value 755 756 # Diagonal elements (second derivatives) 757 for i in range(n): 758 if verbose: 759 vedo.printc(f"Computing Hessian: {i+1}/{n} for diagonal", delay=0) 760 if bounds: 761 lb, ub = bounds[i] 762 else: 763 lb, ub = -np.inf, np.inf 764 765 # Adaptive step size to stay within bounds 766 h_plus = min(epsilon, ub - params[i]) # Max allowed step upward 767 h_minus = min(epsilon, params[i] - lb) # Max allowed step downward 768 h = min(h_plus, h_minus) # Use symmetric step where possible 769 770 # Avoid zero step size (if parameter is at a boundary) 771 if h <= 0: 772 h = epsilon # Fallback to one-sided derivative 773 774 # Compute f(x + h) and f(x - h) 775 params_plus = np.copy(params) 776 params_plus[i] = np.clip(params[i] + h, lb, ub) 777 f_plus = func(params_plus) 778 779 params_minus = np.copy(params) 780 params_minus[i] = np.clip(params[i] - h, lb, ub) 781 f_minus = func(params_minus) 782 783 # Central difference for diagonal 784 hessian[i, i] = (f_plus - 2*f_0 + f_minus) / (h**2) 785 786 # Off-diagonal elements (mixed partial derivatives) 787 for i in range(n): 788 if verbose: 789 print(f"Computing Hessian: {i+1}/{n} for off-diagonal ", end='') 790 for j in range(i + 1, n): 791 if verbose: 792 print(f".", end='') 793 if bounds: 794 lb_i, ub_i = bounds[i] 795 lb_j, ub_j = bounds[j] 796 else: 797 lb_i, ub_i = -np.inf, np.inf 798 lb_j, ub_j = -np.inf, np.inf 799 800 # Step sizes for i and j 801 h_i_plus = min(epsilon, ub_i - params[i]) 802 h_i_minus = min(epsilon, params[i] - lb_i) 803 h_i = min(h_i_plus, h_i_minus) 804 h_i = max(h_i, 1e-10) # Prevent division by zero 805 806 h_j_plus = min(epsilon, ub_j - params[j]) 807 h_j_minus = min(epsilon, params[j] - lb_j) 808 h_j = min(h_j_plus, h_j_minus) 809 h_j = max(h_j, 1e-10) 810 811 # Compute four perturbed points 812 params_pp = np.copy(params) 813 params_pp[i] = np.clip(params[i] + h_i, lb_i, ub_i) 814 params_pp[j] = np.clip(params[j] + h_j, lb_j, ub_j) 815 f_pp = func(params_pp) 816 817 params_pm = np.copy(params) 818 params_pm[i] = np.clip(params[i] + h_i, lb_i, ub_i) 819 params_pm[j] = np.clip(params[j] - h_j, lb_j, ub_j) 820 f_pm = func(params_pm) 821 822 params_mp = np.copy(params) 823 params_mp[i] = np.clip(params[i] - h_i, lb_i, ub_i) 824 params_mp[j] = np.clip(params[j] + h_j, lb_j, ub_j) 825 f_mp = func(params_mp) 826 827 params_mm = np.copy(params) 828 params_mm[i] = np.clip(params[i] - h_i, lb_i, ub_i) 829 params_mm[j] = np.clip(params[j] - h_j, lb_j, ub_j) 830 f_mm = func(params_mm) 831 832 # Central difference for off-diagonal 833 hessian[i, j] = (f_pp - f_pm - f_mp + f_mm) / (4 * h_i * h_j) 834 hessian[j, i] = hessian[i, j] # Symmetric 835 if verbose: 836 print() 837 return hessian 838 839 840########################################################### 841def andrews_curves(M, res=100) -> np.ndarray: 842 """ 843 Computes the [Andrews curves](https://en.wikipedia.org/wiki/Andrews_plot) 844 for the provided data. 845 846 The input array is an array of shape (n,m) where n is the number of 847 features and m is the number of observations. 848 849 Arguments: 850 M : (ndarray) 851 the data matrix (or data vector). 852 res : (int) 853 the resolution (n. of points) of the output curve. 854 855 Example: 856 - [andrews_cluster.py](https://github.com/marcomusy/vedo/blob/master/examples/pyplot/andrews_cluster.py) 857 858 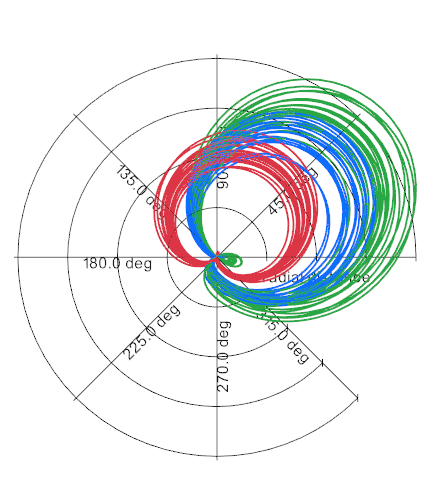 859 """ 860 # Credits: 861 # https://gist.github.com/ryuzakyl/12c221ff0e54d8b1ac171c69ea552c0a 862 M = np.asarray(M) 863 m = int(res + 0.5) 864 865 # getting data vectors 866 X = np.reshape(M, (1, -1)) if len(M.shape) == 1 else M.copy() 867 _rows, n = X.shape 868 869 # andrews curve dimension (n. theta angles) 870 t = np.linspace(-np.pi, np.pi, m) 871 872 # m: range of values for angle theta 873 # n: amount of components of the Fourier expansion 874 A = np.empty((m, n)) 875 876 # setting first column of A 877 A[:, 0] = [1/np.sqrt(2)] * m 878 879 # filling columns of A 880 for i in range(1, n): 881 # computing the scaling coefficient for angle theta 882 c = np.ceil(i / 2) 883 # computing i-th column of matrix A 884 col = np.sin(c * t) if i % 2 == 1 else np.cos(c * t) 885 # setting column in matrix A 886 A[:, i] = col[:] 887 888 # computing Andrews curves for provided data 889 andrew_curves = np.dot(A, X.T).T 890 891 # returning the Andrews Curves (raveling if needed) 892 return np.ravel(andrew_curves) if andrew_curves.shape[0] == 1 else andrew_curves 893 894 895########################################################### 896def numpy2vtk(arr, dtype=None, deep=True, name=""): 897 """ 898 Convert a numpy array into a `vtkDataArray`. 899 Use `dtype='id'` for `vtkIdTypeArray` objects. 900 """ 901 # https://github.com/Kitware/VTK/blob/master/Wrapping/Python/vtkmodules/util/numpy_support.py 902 if arr is None: 903 return None 904 905 arr = np.ascontiguousarray(arr) 906 907 if dtype == "id": 908 if vtki.vtkIdTypeArray().GetDataTypeSize() != 4: 909 ast = np.int64 910 else: 911 ast = np.int32 912 varr = numpy_to_vtkIdTypeArray(arr.astype(ast), deep=deep) 913 elif dtype: 914 varr = numpy_to_vtk(arr.astype(dtype), deep=deep) 915 else: 916 # let numpy_to_vtk() decide what is best type based on arr type 917 if arr.dtype == np.bool_: 918 arr = arr.astype(np.uint8) 919 varr = numpy_to_vtk(arr, deep=deep) 920 921 if name: 922 varr.SetName(name) 923 return varr 924 925def vtk2numpy(varr): 926 """Convert a `vtkDataArray`, `vtkIdList` or `vtTransform` into a numpy array.""" 927 if varr is None: 928 return np.array([]) 929 if isinstance(varr, vtki.vtkIdList): 930 return np.array([varr.GetId(i) for i in range(varr.GetNumberOfIds())]) 931 elif isinstance(varr, vtki.vtkBitArray): 932 carr = vtki.vtkCharArray() 933 carr.DeepCopy(varr) 934 varr = carr 935 elif isinstance(varr, vtki.vtkHomogeneousTransform): 936 try: 937 varr = varr.GetMatrix() 938 except AttributeError: 939 pass 940 M = [[varr.GetElement(i, j) for j in range(4)] for i in range(4)] 941 return np.array(M) 942 return vtk_to_numpy(varr) 943 944 945def make3d(pts) -> np.ndarray: 946 """ 947 Make an array which might be 2D to 3D. 948 949 Array can also be in the form `[allx, ally, allz]`. 950 """ 951 if pts is None: 952 return np.array([]) 953 pts = np.asarray(pts) 954 955 if pts.dtype == "object": 956 raise ValueError("Cannot form a valid numpy array, input may be non-homogenous") 957 958 if pts.size == 0: # empty list 959 return pts 960 961 if pts.ndim == 1: 962 if pts.shape[0] == 2: 963 return np.hstack([pts, [0]]).astype(pts.dtype) 964 elif pts.shape[0] == 3: 965 return pts 966 else: 967 raise ValueError 968 969 if pts.shape[1] == 3: 970 return pts 971 972 # if 2 <= pts.shape[0] <= 3 and pts.shape[1] > 3: 973 # pts = pts.T 974 975 if pts.shape[1] == 2: 976 return np.c_[pts, np.zeros(pts.shape[0], dtype=pts.dtype)] 977 978 if pts.shape[1] != 3: 979 raise ValueError(f"input shape is not supported: {pts.shape}") 980 return pts 981 982 983def geometry(obj, extent=None) -> "vedo.Mesh": 984 """ 985 Apply the `vtkGeometryFilter` to the input object. 986 This is a general-purpose filter to extract geometry (and associated data) 987 from any type of dataset. 988 This filter also may be used to convert any type of data to polygonal type. 989 The conversion process may be less than satisfactory for some 3D datasets. 990 For example, this filter will extract the outer surface of a volume 991 or structured grid dataset. 992 993 Returns a `vedo.Mesh` object. 994 995 Set `extent` as the `[xmin,xmax, ymin,ymax, zmin,zmax]` bounding box to clip data. 996 """ 997 gf = vtki.new("GeometryFilter") 998 gf.SetInputData(obj) 999 if extent is not None: 1000 gf.SetExtent(extent) 1001 gf.Update() 1002 return vedo.Mesh(gf.GetOutput()) 1003 1004 1005def buildPolyData(vertices, faces=None, lines=None, strips=None, index_offset=0) -> vtki.vtkPolyData: 1006 """ 1007 Build a `vtkPolyData` object from a list of vertices 1008 where faces represents the connectivity of the polygonal mesh. 1009 Lines and triangle strips can also be specified. 1010 1011 E.g. : 1012 - `vertices=[[x1,y1,z1],[x2,y2,z2], ...]` 1013 - `faces=[[0,1,2], [1,2,3], ...]` 1014 - `lines=[[0,1], [1,2,3,4], ...]` 1015 - `strips=[[0,1,2,3,4,5], [2,3,9,7,4], ...]` 1016 1017 A flat list of faces can be passed as `faces=[3, 0,1,2, 4, 1,2,3,4, ...]`. 1018 For lines use `lines=[2, 0,1, 4, 1,2,3,4, ...]`. 1019 1020 Use `index_offset=1` if face numbering starts from 1 instead of 0. 1021 """ 1022 if is_sequence(faces) and len(faces) == 0: 1023 faces=None 1024 if is_sequence(lines) and len(lines) == 0: 1025 lines=None 1026 if is_sequence(strips) and len(strips) == 0: 1027 strips=None 1028 1029 poly = vtki.vtkPolyData() 1030 1031 if len(vertices) == 0: 1032 return poly 1033 1034 vertices = make3d(vertices) 1035 source_points = vtki.vtkPoints() 1036 if vedo.settings.force_single_precision_points: 1037 source_points.SetData(numpy2vtk(vertices, dtype=np.float32)) 1038 else: 1039 source_points.SetData(numpy2vtk(vertices)) 1040 poly.SetPoints(source_points) 1041 1042 if lines is not None: 1043 # Create a cell array to store the lines in and add the lines to it 1044 linesarr = vtki.vtkCellArray() 1045 if is_sequence(lines[0]): # assume format [(id0,id1),..] 1046 for iline in lines: 1047 for i in range(0, len(iline) - 1): 1048 i1, i2 = iline[i], iline[i + 1] 1049 if i1 != i2: 1050 vline = vtki.vtkLine() 1051 vline.GetPointIds().SetId(0, i1) 1052 vline.GetPointIds().SetId(1, i2) 1053 linesarr.InsertNextCell(vline) 1054 else: # assume format [id0,id1,...] 1055 # print("buildPolyData: assuming lines format [id0,id1,...]", lines) 1056 # TODO CORRECT THIS CASE, MUST BE [2, id0,id1,...] 1057 for i in range(0, len(lines) - 1): 1058 vline = vtki.vtkLine() 1059 vline.GetPointIds().SetId(0, lines[i]) 1060 vline.GetPointIds().SetId(1, lines[i + 1]) 1061 linesarr.InsertNextCell(vline) 1062 poly.SetLines(linesarr) 1063 1064 if faces is not None: 1065 1066 source_polygons = vtki.vtkCellArray() 1067 1068 if isinstance(faces, np.ndarray) or not is_ragged(faces): 1069 ##### all faces are composed of equal nr of vtxs, FAST 1070 faces = np.asarray(faces) 1071 if vtki.vtkIdTypeArray().GetDataTypeSize() != 4: 1072 ast = np.int64 1073 else: 1074 ast = np.int32 1075 1076 if faces.ndim > 1: 1077 nf, nc = faces.shape 1078 hs = np.hstack((np.zeros(nf)[:, None] + nc, faces)) 1079 else: 1080 nf = faces.shape[0] 1081 hs = faces 1082 arr = numpy_to_vtkIdTypeArray(hs.astype(ast).ravel(), deep=True) 1083 source_polygons.SetCells(nf, arr) 1084 1085 else: 1086 ############################# manually add faces, SLOW 1087 for f in faces: 1088 n = len(f) 1089 1090 if n == 3: 1091 tri = vtki.vtkTriangle() 1092 pids = tri.GetPointIds() 1093 for i in range(3): 1094 pids.SetId(i, f[i] - index_offset) 1095 source_polygons.InsertNextCell(tri) 1096 1097 else: 1098 ele = vtki.vtkPolygon() 1099 pids = ele.GetPointIds() 1100 pids.SetNumberOfIds(n) 1101 for i in range(n): 1102 pids.SetId(i, f[i] - index_offset) 1103 source_polygons.InsertNextCell(ele) 1104 1105 poly.SetPolys(source_polygons) 1106 1107 if strips is not None: 1108 tscells = vtki.vtkCellArray() 1109 for strip in strips: 1110 # create a triangle strip 1111 # https://vtk.org/doc/nightly/html/classvtkTriangleStrip.html 1112 n = len(strip) 1113 tstrip = vtki.vtkTriangleStrip() 1114 tstrip_ids = tstrip.GetPointIds() 1115 tstrip_ids.SetNumberOfIds(n) 1116 for i in range(n): 1117 tstrip_ids.SetId(i, strip[i] - index_offset) 1118 tscells.InsertNextCell(tstrip) 1119 poly.SetStrips(tscells) 1120 1121 if faces is None and lines is None and strips is None: 1122 source_vertices = vtki.vtkCellArray() 1123 for i in range(len(vertices)): 1124 source_vertices.InsertNextCell(1) 1125 source_vertices.InsertCellPoint(i) 1126 poly.SetVerts(source_vertices) 1127 1128 # print("buildPolyData \n", 1129 # poly.GetNumberOfPoints(), 1130 # poly.GetNumberOfCells(), # grand total 1131 # poly.GetNumberOfLines(), 1132 # poly.GetNumberOfPolys(), 1133 # poly.GetNumberOfStrips(), 1134 # poly.GetNumberOfVerts(), 1135 # ) 1136 return poly 1137 1138 1139############################################################################## 1140def get_font_path(font: str) -> str: 1141 """Internal use.""" 1142 if font in vedo.settings.font_parameters.keys(): 1143 if vedo.settings.font_parameters[font]["islocal"]: 1144 fl = os.path.join(vedo.fonts_path, f"{font}.ttf") 1145 else: 1146 try: 1147 fl = vedo.file_io.download(f"https://vedo.embl.es/fonts/{font}.ttf", verbose=False) 1148 except: 1149 vedo.logger.warning(f"Could not download https://vedo.embl.es/fonts/{font}.ttf") 1150 fl = os.path.join(vedo.fonts_path, "Normografo.ttf") 1151 else: 1152 if font.startswith("https://"): 1153 fl = vedo.file_io.download(font, verbose=False) 1154 elif os.path.isfile(font): 1155 fl = font # assume user is passing a valid file 1156 else: 1157 if font.endswith(".ttf"): 1158 vedo.logger.error( 1159 f"Could not set font file {font}" 1160 f"-> using default: {vedo.settings.default_font}" 1161 ) 1162 else: 1163 vedo.settings.default_font = "Normografo" 1164 vedo.logger.error( 1165 f"Could not set font name {font}" 1166 f" -> using default: Normografo\n" 1167 f"Check out https://vedo.embl.es/fonts for additional fonts\n" 1168 f"Type 'vedo -r fonts' to see available fonts" 1169 ) 1170 fl = get_font_path(vedo.settings.default_font) 1171 return fl 1172 1173 1174def is_sequence(arg) -> bool: 1175 """Check if the input is iterable.""" 1176 if hasattr(arg, "strip"): 1177 return False 1178 if hasattr(arg, "__getslice__"): 1179 return True 1180 if hasattr(arg, "__iter__"): 1181 return True 1182 return False 1183 1184 1185def is_ragged(arr, deep=False) -> bool: 1186 """ 1187 A ragged or inhomogeneous array in Python is an array 1188 with arrays of different lengths as its elements. 1189 To check if an array is ragged, we iterate through the elements 1190 and check if their lengths are the same. 1191 1192 Example: 1193 ```python 1194 arr = [[1, 2, 3], [[4, 5], [6], 1], [7, 8, 9]] 1195 print(is_ragged(arr, deep=True)) # output: True 1196 ``` 1197 """ 1198 n = len(arr) 1199 if n == 0: 1200 return False 1201 if is_sequence(arr[0]): 1202 length = len(arr[0]) 1203 for i in range(1, n): 1204 if len(arr[i]) != length or (deep and is_ragged(arr[i])): 1205 return True 1206 return False 1207 return False 1208 1209 1210def flatten(list_to_flatten) -> list: 1211 """Flatten out a list.""" 1212 1213 def _genflatten(lst): 1214 for elem in lst: 1215 if isinstance(elem, (list, tuple)): 1216 for x in flatten(elem): 1217 yield x 1218 else: 1219 yield elem 1220 1221 return list(_genflatten(list_to_flatten)) 1222 1223 1224def humansort(alist) -> list: 1225 """ 1226 Sort in place a given list the way humans expect. 1227 1228 E.g. `['file11', 'file1'] -> ['file1', 'file11']` 1229 1230 .. warning:: input list is modified in-place by this function. 1231 """ 1232 import re 1233 1234 def alphanum_key(s): 1235 # Turn a string into a list of string and number chunks. 1236 # e.g. "z23a" -> ["z", 23, "a"] 1237 def tryint(s): 1238 if s.isdigit(): 1239 return int(s) 1240 return s 1241 1242 return [tryint(c) for c in re.split("([0-9]+)", s)] 1243 1244 alist.sort(key=alphanum_key) 1245 return alist # NB: input list is modified 1246 1247 1248def sort_by_column(arr, nth, invert=False) -> np.ndarray: 1249 """Sort a numpy array by its `n-th` column.""" 1250 arr = np.asarray(arr) 1251 arr = arr[arr[:, nth].argsort()] 1252 if invert: 1253 return np.flip(arr, axis=0) 1254 return arr 1255 1256 1257def point_in_triangle(p, p1, p2, p3) -> Union[bool, None]: 1258 """ 1259 Return True if a point is inside (or above/below) 1260 a triangle defined by 3 points in space. 1261 """ 1262 p1 = np.array(p1) 1263 u = p2 - p1 1264 v = p3 - p1 1265 n = np.cross(u, v) 1266 w = p - p1 1267 ln = np.dot(n, n) 1268 if not ln: 1269 return None # degenerate triangle 1270 gamma = (np.dot(np.cross(u, w), n)) / ln 1271 if 0 < gamma < 1: 1272 beta = (np.dot(np.cross(w, v), n)) / ln 1273 if 0 < beta < 1: 1274 alpha = 1 - gamma - beta 1275 if 0 < alpha < 1: 1276 return True 1277 return False 1278 1279 1280def intersection_ray_triangle(P0, P1, V0, V1, V2) -> Union[bool, None, np.ndarray]: 1281 """ 1282 Fast intersection between a directional ray defined by `P0,P1` 1283 and triangle `V0, V1, V2`. 1284 1285 Returns the intersection point or 1286 - `None` if triangle is degenerate, or ray is parallel to triangle plane. 1287 - `False` if no intersection, or ray direction points away from triangle. 1288 """ 1289 # Credits: http://geomalgorithms.com/a06-_intersect-2.html 1290 # Get triangle edge vectors and plane normal 1291 # todo : this is slow should check 1292 # https://vtk.org/doc/nightly/html/classvtkCell.html 1293 V0 = np.asarray(V0, dtype=float) 1294 P0 = np.asarray(P0, dtype=float) 1295 u = V1 - V0 1296 v = V2 - V0 1297 n = np.cross(u, v) 1298 if not np.abs(v).sum(): # triangle is degenerate 1299 return None # do not deal with this case 1300 1301 rd = P1 - P0 # ray direction vector 1302 w0 = P0 - V0 1303 a = -np.dot(n, w0) 1304 b = np.dot(n, rd) 1305 if not b: # ray is parallel to triangle plane 1306 return None 1307 1308 # Get intersect point of ray with triangle plane 1309 r = a / b 1310 if r < 0.0: # ray goes away from triangle 1311 return False # => no intersect 1312 1313 # Gor a segment, also test if (r > 1.0) => no intersect 1314 I = P0 + r * rd # intersect point of ray and plane 1315 1316 # is I inside T? 1317 uu = np.dot(u, u) 1318 uv = np.dot(u, v) 1319 vv = np.dot(v, v) 1320 w = I - V0 1321 wu = np.dot(w, u) 1322 wv = np.dot(w, v) 1323 D = uv * uv - uu * vv 1324 1325 # Get and test parametric coords 1326 s = (uv * wv - vv * wu) / D 1327 if s < 0.0 or s > 1.0: # I is outside T 1328 return False 1329 t = (uv * wu - uu * wv) / D 1330 if t < 0.0 or (s + t) > 1.0: # I is outside T 1331 return False 1332 return I # I is in T 1333 1334 1335def triangle_solver(**input_dict): 1336 """ 1337 Solve a triangle from any 3 known elements. 1338 (Note that there might be more than one solution or none). 1339 Angles are in radians. 1340 1341 Example: 1342 ```python 1343 print(triangle_solver(a=3, b=4, c=5)) 1344 print(triangle_solver(a=3, ac=0.9273, ab=1.5716)) 1345 print(triangle_solver(a=3, b=4, ab=1.5716)) 1346 print(triangle_solver(b=4, bc=.64, ab=1.5716)) 1347 print(triangle_solver(c=5, ac=.9273, bc=0.6435)) 1348 print(triangle_solver(a=3, c=5, bc=0.6435)) 1349 print(triangle_solver(b=4, c=5, ac=0.927)) 1350 ``` 1351 """ 1352 a = input_dict.get("a") 1353 b = input_dict.get("b") 1354 c = input_dict.get("c") 1355 ab = input_dict.get("ab") 1356 bc = input_dict.get("bc") 1357 ac = input_dict.get("ac") 1358 1359 if ab and bc: 1360 ac = np.pi - bc - ab 1361 elif bc and ac: 1362 ab = np.pi - bc - ac 1363 elif ab and ac: 1364 bc = np.pi - ab - ac 1365 1366 if a is not None and b is not None and c is not None: 1367 ab = np.arccos((a ** 2 + b ** 2 - c ** 2) / (2 * a * b)) 1368 sinab = np.sin(ab) 1369 ac = np.arcsin(a / c * sinab) 1370 bc = np.arcsin(b / c * sinab) 1371 1372 elif a is not None and b is not None and ab is not None: 1373 c = np.sqrt(a ** 2 + b ** 2 - 2 * a * b * np.cos(ab)) 1374 sinab = np.sin(ab) 1375 ac = np.arcsin(a / c * sinab) 1376 bc = np.arcsin(b / c * sinab) 1377 1378 elif a is not None and ac is not None and ab is not None: 1379 h = a * np.sin(ac) 1380 b = h / np.sin(bc) 1381 c = b * np.cos(bc) + a * np.cos(ac) 1382 1383 elif b is not None and bc is not None and ab is not None: 1384 h = b * np.sin(bc) 1385 a = h / np.sin(ac) 1386 c = np.sqrt(a * a + b * b) 1387 1388 elif c is not None and ac is not None and bc is not None: 1389 h = c * np.sin(bc) 1390 b1 = c * np.cos(bc) 1391 b2 = h / np.tan(ab) 1392 b = b1 + b2 1393 a = np.sqrt(b2 * b2 + h * h) 1394 1395 elif a is not None and c is not None and bc is not None: 1396 # double solution 1397 h = c * np.sin(bc) 1398 k = np.sqrt(a * a - h * h) 1399 omega = np.arcsin(k / a) 1400 cosbc = np.cos(bc) 1401 b = c * cosbc - k 1402 phi = np.pi / 2 - bc - omega 1403 ac = phi 1404 ab = np.pi - ac - bc 1405 if k: 1406 b2 = c * cosbc + k 1407 ac2 = phi + 2 * omega 1408 ab2 = np.pi - ac2 - bc 1409 return [ 1410 {"a": a, "b": b, "c": c, "ab": ab, "bc": bc, "ac": ac}, 1411 {"a": a, "b": b2, "c": c, "ab": ab2, "bc": bc, "ac": ac2}, 1412 ] 1413 1414 elif b is not None and c is not None and ac is not None: 1415 # double solution 1416 h = c * np.sin(ac) 1417 k = np.sqrt(b * b - h * h) 1418 omega = np.arcsin(k / b) 1419 cosac = np.cos(ac) 1420 a = c * cosac - k 1421 phi = np.pi / 2 - ac - omega 1422 bc = phi 1423 ab = np.pi - bc - ac 1424 if k: 1425 a2 = c * cosac + k 1426 bc2 = phi + 2 * omega 1427 ab2 = np.pi - ac - bc2 1428 return [ 1429 {"a": a, "b": b, "c": c, "ab": ab, "bc": bc, "ac": ac}, 1430 {"a": a2, "b": b, "c": c, "ab": ab2, "bc": bc2, "ac": ac}, 1431 ] 1432 1433 else: 1434 vedo.logger.error(f"Case {input_dict} is not supported.") 1435 return [] 1436 1437 return [{"a": a, "b": b, "c": c, "ab": ab, "bc": bc, "ac": ac}] 1438 1439 1440############################################################################# 1441def circle_from_3points(p1, p2, p3) -> np.ndarray: 1442 """ 1443 Find the center and radius of a circle given 3 points in 3D space. 1444 1445 Returns the center of the circle. 1446 1447 Example: 1448 ```python 1449 from vedo.utils import mag, circle_from_3points 1450 p1 = [0,1,1] 1451 p2 = [3,0,1] 1452 p3 = [1,2,0] 1453 c = circle_from_3points(p1, p2, p3) 1454 print(mag(c-p1), mag(c-p2), mag(c-p3)) 1455 ``` 1456 """ 1457 p1 = np.asarray(p1) 1458 p2 = np.asarray(p2) 1459 p3 = np.asarray(p3) 1460 v1 = p2 - p1 1461 v2 = p3 - p1 1462 v11 = np.dot(v1, v1) 1463 v22 = np.dot(v2, v2) 1464 v12 = np.dot(v1, v2) 1465 f = 1.0 / (2 * (v11 * v22 - v12 * v12)) 1466 k1 = f * v22 * (v11-v12) 1467 k2 = f * v11 * (v22-v12) 1468 return p1 + k1 * v1 + k2 * v2 1469 1470def point_line_distance(p, p1, p2) -> float: 1471 """ 1472 Compute the distance of a point to a line (not the segment) 1473 defined by `p1` and `p2`. 1474 """ 1475 return np.sqrt(vtki.vtkLine.DistanceToLine(p, p1, p2)) 1476 1477def line_line_distance(p1, p2, q1, q2) -> Tuple[float, np.ndarray, np.ndarray, float, float]: 1478 """ 1479 Compute the distance of a line to a line (not the segment) 1480 defined by `p1` and `p2` and `q1` and `q2`. 1481 1482 Returns the distance, 1483 the closest point on line 1, the closest point on line 2. 1484 Their parametric coords (-inf <= t0, t1 <= inf) are also returned. 1485 """ 1486 closest_pt1: MutableSequence[float] = [0,0,0] 1487 closest_pt2: MutableSequence[float] = [0,0,0] 1488 t1, t2 = 0.0, 0.0 1489 d = vtki.vtkLine.DistanceBetweenLines( 1490 p1, p2, q1, q2, closest_pt1, closest_pt2, t1, t2) 1491 return np.sqrt(d), closest_pt1, closest_pt2, t1, t2 1492 1493def segment_segment_distance(p1, p2, q1, q2): 1494 """ 1495 Compute the distance of a segment to a segment 1496 defined by `p1` and `p2` and `q1` and `q2`. 1497 1498 Returns the distance, 1499 the closest point on line 1, the closest point on line 2. 1500 Their parametric coords (-inf <= t0, t1 <= inf) are also returned. 1501 """ 1502 closest_pt1 = [0,0,0] 1503 closest_pt2 = [0,0,0] 1504 t1, t2 = 0.0, 0.0 1505 d = vtki.vtkLine.DistanceBetweenLineSegments( 1506 p1, p2, q1, q2, closest_pt1, closest_pt2, t1, t2) 1507 return np.sqrt(d), closest_pt1, closest_pt2, t1, t2 1508 1509 1510def closest(point, points, n=1, return_ids=False, use_tree=False): 1511 """ 1512 Returns the distances and the closest point(s) to the given set of points. 1513 Needs `scipy.spatial` library. 1514 1515 Arguments: 1516 n : (int) 1517 the nr of closest points to return 1518 return_ids : (bool) 1519 return the ids instead of the points coordinates 1520 use_tree : (bool) 1521 build a `scipy.spatial.KDTree`. 1522 An already existing one can be passed to avoid rebuilding. 1523 """ 1524 from scipy.spatial import distance, KDTree 1525 1526 points = np.asarray(points) 1527 if n == 1: 1528 dists = distance.cdist([point], points) 1529 closest_idx = np.argmin(dists) 1530 else: 1531 if use_tree: 1532 if isinstance(use_tree, KDTree): # reuse 1533 tree = use_tree 1534 else: 1535 tree = KDTree(points) 1536 dists, closest_idx = tree.query([point], k=n) 1537 closest_idx = closest_idx[0] 1538 else: 1539 dists = distance.cdist([point], points) 1540 closest_idx = np.argsort(dists)[0][:n] 1541 if return_ids: 1542 return dists, closest_idx 1543 else: 1544 return dists, points[closest_idx] 1545 1546 1547############################################################################# 1548def lin_interpolate(x, rangeX, rangeY): 1549 """ 1550 Interpolate linearly the variable `x` in `rangeX` onto the new `rangeY`. 1551 If `x` is a 3D vector the linear weight is the distance to the two 3D `rangeX` vectors. 1552 1553 E.g. if `x` runs in `rangeX=[x0,x1]` and I want it to run in `rangeY=[y0,y1]` then 1554 1555 `y = lin_interpolate(x, rangeX, rangeY)` will interpolate `x` onto `rangeY`. 1556 1557 Examples: 1558 - [lin_interpolate.py](https://github.com/marcomusy/vedo/tree/master/examples/basic/lin_interpolate.py) 1559 1560 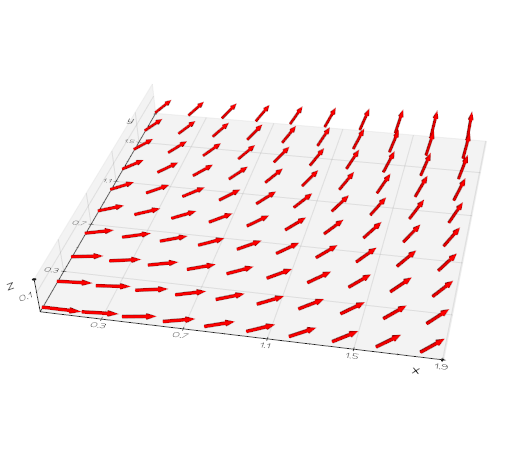 1561 """ 1562 if is_sequence(x): 1563 x = np.asarray(x) 1564 x0, x1 = np.asarray(rangeX) 1565 y0, y1 = np.asarray(rangeY) 1566 dx = x1 - x0 1567 dxn = np.linalg.norm(dx) 1568 if not dxn: 1569 return y0 1570 s = np.linalg.norm(x - x0) / dxn 1571 t = np.linalg.norm(x - x1) / dxn 1572 st = s + t 1573 out = y0 * (t / st) + y1 * (s / st) 1574 1575 else: # faster 1576 1577 x0 = rangeX[0] 1578 dx = rangeX[1] - x0 1579 if not dx: 1580 return rangeY[0] 1581 s = (x - x0) / dx 1582 out = rangeY[0] * (1 - s) + rangeY[1] * s 1583 return out 1584 1585 1586def get_uv(p, x, v): 1587 """ 1588 Obtain the texture uv-coords of a point p belonging to a face that has point 1589 coordinates (x0, x1, x2) with the corresponding uv-coordinates v=(v0, v1, v2). 1590 All p and x0,x1,x2 are 3D-vectors, while v are their 2D uv-coordinates. 1591 1592 Example: 1593 ```python 1594 from vedo import * 1595 1596 pic = Image(dataurl+"coloured_cube_faces.jpg") 1597 cb = Mesh(dataurl+"coloured_cube.obj").lighting("off").texture(pic) 1598 1599 cbpts = cb.points 1600 faces = cb.cells 1601 uv = cb.pointdata["Material"] 1602 1603 pt = [-0.2, 0.75, 2] 1604 pr = cb.closest_point(pt) 1605 1606 idface = cb.closest_point(pt, return_cell_id=True) 1607 idpts = faces[idface] 1608 uv_face = uv[idpts] 1609 1610 uv_pr = utils.get_uv(pr, cbpts[idpts], uv_face) 1611 print("interpolated uv =", uv_pr) 1612 1613 sx, sy = pic.dimensions() 1614 i_interp_uv = uv_pr * [sy, sx] 1615 ix, iy = i_interp_uv.astype(int) 1616 mpic = pic.tomesh() 1617 rgba = mpic.pointdata["RGBA"].reshape(sy, sx, 3) 1618 print("color =", rgba[ix, iy]) 1619 1620 show( 1621 [[cb, Point(pr), cb.labels("Material")], 1622 [pic, Point(i_interp_uv)]], 1623 N=2, axes=1, sharecam=False, 1624 ).close() 1625 ``` 1626 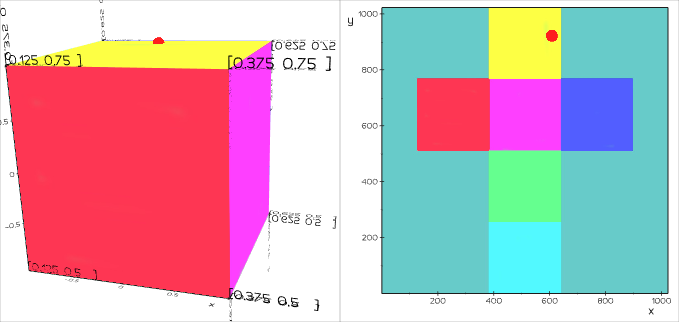 1627 """ 1628 # Vector vp=p-x0 is representable as alpha*s + beta*t, 1629 # where s = x1-x0 and t = x2-x0, in matrix form 1630 # vp = [alpha, beta] . matrix(s,t) 1631 # M = matrix(s,t) is 2x3 matrix, so (alpha, beta) can be found by 1632 # inverting any of its minor A with non-zero determinant. 1633 # Once found, uv-coords of p are vt0 + alpha (vt1-v0) + beta (vt2-v0) 1634 1635 p = np.asarray(p) 1636 x0, x1, x2 = np.asarray(x)[:3] 1637 vt0, vt1, vt2 = np.asarray(v)[:3] 1638 1639 s = x1 - x0 1640 t = x2 - x0 1641 vs = vt1 - vt0 1642 vt = vt2 - vt0 1643 vp = p - x0 1644 1645 # finding a minor with independent rows 1646 M = np.matrix([s, t]) 1647 mnr = [0, 1] 1648 A = M[:, mnr] 1649 if np.abs(np.linalg.det(A)) < 0.000001: 1650 mnr = [0, 2] 1651 A = M[:, mnr] 1652 if np.abs(np.linalg.det(A)) < 0.000001: 1653 mnr = [1, 2] 1654 A = M[:, mnr] 1655 Ainv = np.linalg.inv(A) 1656 alpha_beta = vp[mnr].dot(Ainv) # [alpha, beta] 1657 return np.asarray(vt0 + alpha_beta.dot(np.matrix([vs, vt])))[0] 1658 1659 1660def vector(x, y=None, z=0.0, dtype=np.float64) -> np.ndarray: 1661 """ 1662 Return a 3D numpy array representing a vector. 1663 1664 If `y` is `None`, assume input is already in the form `[x,y,z]`. 1665 """ 1666 if y is None: # assume x is already [x,y,z] 1667 return np.asarray(x, dtype=dtype) 1668 return np.array([x, y, z], dtype=dtype) 1669 1670 1671def versor(x, y=None, z=0.0, dtype=np.float64) -> np.ndarray: 1672 """Return the unit vector. Input can be a list of vectors.""" 1673 v = vector(x, y, z, dtype) 1674 if isinstance(v[0], np.ndarray): 1675 return np.divide(v, mag(v)[:, None]) 1676 return v / mag(v) 1677 1678 1679def mag(v): 1680 """Get the magnitude of a vector or array of vectors.""" 1681 v = np.asarray(v) 1682 if v.ndim == 1: 1683 return np.linalg.norm(v) 1684 return np.linalg.norm(v, axis=1) 1685 1686 1687def mag2(v) -> np.ndarray: 1688 """Get the squared magnitude of a vector or array of vectors.""" 1689 v = np.asarray(v) 1690 if v.ndim == 1: 1691 return np.square(v).sum() 1692 return np.square(v).sum(axis=1) 1693 1694 1695def is_integer(n) -> bool: 1696 """Check if input is an integer.""" 1697 try: 1698 float(n) 1699 except (ValueError, TypeError): 1700 return False 1701 else: 1702 return float(n).is_integer() 1703 1704 1705def is_number(n) -> bool: 1706 """Check if input is a number""" 1707 try: 1708 float(n) 1709 return True 1710 except (ValueError, TypeError): 1711 return False 1712 1713 1714def round_to_digit(x, p) -> float: 1715 """Round a real number to the specified number of significant digits.""" 1716 if not x: 1717 return 0 1718 r = np.round(x, p - int(np.floor(np.log10(abs(x)))) - 1) 1719 if int(r) == r: 1720 return int(r) 1721 return r 1722 1723 1724def pack_spheres(bounds, radius) -> np.ndarray: 1725 """ 1726 Packing spheres into a bounding box. 1727 Returns a numpy array of sphere centers. 1728 """ 1729 h = 0.8164965 / 2 1730 d = 0.8660254 1731 a = 0.288675135 1732 1733 if is_sequence(bounds): 1734 x0, x1, y0, y1, z0, z1 = bounds 1735 else: 1736 x0, x1, y0, y1, z0, z1 = bounds.bounds() 1737 1738 x = np.arange(x0, x1, radius) 1739 nul = np.zeros_like(x) 1740 nz = int((z1 - z0) / radius / h / 2 + 1.5) 1741 ny = int((y1 - y0) / radius / d + 1.5) 1742 1743 pts = [] 1744 for iz in range(nz): 1745 z = z0 + nul + iz * h * radius 1746 dx, dy, dz = [radius * 0.5, radius * a, iz * h * radius] 1747 for iy in range(ny): 1748 y = y0 + nul + iy * d * radius 1749 if iy % 2: 1750 xs = x 1751 else: 1752 xs = x + radius * 0.5 1753 if iz % 2: 1754 p = np.c_[xs, y, z] + [dx, dy, dz] 1755 else: 1756 p = np.c_[xs, y, z] + [0, 0, dz] 1757 pts += p.tolist() 1758 return np.array(pts) 1759 1760 1761def precision(x, p: int, vrange=None, delimiter="e") -> str: 1762 """ 1763 Returns a string representation of `x` formatted to precision `p`. 1764 1765 Set `vrange` to the range in which x exists (to snap x to '0' if below precision). 1766 """ 1767 # Based on the webkit javascript implementation 1768 # `from here <https://code.google.com/p/webkit-mirror/source/browse/JavaScriptCore/kjs/number_object.cpp>`_, 1769 # and implemented by `randlet <https://github.com/randlet/to-precision>`_. 1770 # Modified for vedo by M.Musy 2020 1771 1772 if isinstance(x, str): # do nothing 1773 return x 1774 1775 if is_sequence(x): 1776 out = "(" 1777 nn = len(x) - 1 1778 for i, ix in enumerate(x): 1779 1780 try: 1781 if np.isnan(ix): 1782 return "NaN" 1783 except: 1784 # cannot handle list of list 1785 continue 1786 1787 out += precision(ix, p) 1788 if i < nn: 1789 out += ", " 1790 return out + ")" ############ <-- 1791 1792 try: 1793 if np.isnan(x): 1794 return "NaN" 1795 except TypeError: 1796 return "NaN" 1797 1798 x = float(x) 1799 1800 if x == 0.0 or (vrange is not None and abs(x) < vrange / pow(10, p)): 1801 return "0" 1802 1803 out = [] 1804 if x < 0: 1805 out.append("-") 1806 x = -x 1807 1808 e = int(np.log10(x)) 1809 # tens = np.power(10, e - p + 1) 1810 tens = 10 ** (e - p + 1) 1811 n = np.floor(x / tens) 1812 1813 # if n < np.power(10, p - 1): 1814 if n < 10 ** (p - 1): 1815 e = e - 1 1816 # tens = np.power(10, e - p + 1) 1817 tens = 10 ** (e - p + 1) 1818 n = np.floor(x / tens) 1819 1820 if abs((n + 1.0) * tens - x) <= abs(n * tens - x): 1821 n = n + 1 1822 1823 # if n >= np.power(10, p): 1824 if n >= 10 ** p: 1825 n = n / 10.0 1826 e = e + 1 1827 1828 m = "%.*g" % (p, n) 1829 if e < -2 or e >= p: 1830 out.append(m[0]) 1831 if p > 1: 1832 out.append(".") 1833 out.extend(m[1:p]) 1834 out.append(delimiter) 1835 if e > 0: 1836 out.append("+") 1837 out.append(str(e)) 1838 elif e == (p - 1): 1839 out.append(m) 1840 elif e >= 0: 1841 out.append(m[: e + 1]) 1842 if e + 1 < len(m): 1843 out.append(".") 1844 out.extend(m[e + 1 :]) 1845 else: 1846 out.append("0.") 1847 out.extend(["0"] * -(e + 1)) 1848 out.append(m) 1849 return "".join(out) 1850 1851 1852################################################################################## 1853def grep(filename: str, tag: str, column=None, first_occurrence_only=False) -> list: 1854 """Greps the line in a file that starts with a specific `tag` string inside the file.""" 1855 import re 1856 1857 with open(str(filename), "r", encoding="UTF-8") as afile: 1858 content = [] 1859 for line in afile: 1860 if re.search(tag, line): 1861 c = line.split() 1862 c[-1] = c[-1].replace("\n", "") 1863 if column is not None: 1864 c = c[column] 1865 content.append(c) 1866 if first_occurrence_only: 1867 break 1868 return content 1869 1870def parse_pattern(query, strings_to_parse) -> list: 1871 """ 1872 Parse a pattern query to a list of strings. 1873 The query string can contain wildcards like * and ?. 1874 1875 Arguments: 1876 query : (str) 1877 the query to parse 1878 strings_to_parse : (str/list) 1879 the string or list of strings to parse 1880 1881 Returns: 1882 a list of booleans, one for each string in strings_to_parse 1883 1884 Example: 1885 >>> query = r'*Sphere 1?3*' 1886 >>> strings = ["Sphere 143 red", "Sphere 13 red", "Sphere 123", "ASphere 173"] 1887 >>> parse_pattern(query, strings) 1888 [True, True, False, False] 1889 """ 1890 from re import findall as re_findall 1891 if not isinstance(query, str): 1892 return [False] 1893 1894 if not is_sequence(strings_to_parse): 1895 strings_to_parse = [strings_to_parse] 1896 1897 outs = [] 1898 for sp in strings_to_parse: 1899 if not isinstance(sp, str): 1900 outs.append(False) 1901 continue 1902 1903 s = query 1904 if s.startswith("*"): 1905 s = s[1:] 1906 else: 1907 s = "^" + s 1908 1909 t = "" 1910 if not s.endswith("*"): 1911 t = "$" 1912 else: 1913 s = s[:-1] 1914 1915 pattern = s.replace('?', r'\w').replace(' ', r'\s').replace("*", r"\w+") + t 1916 1917 # Search for the pattern in the input string 1918 match = re_findall(pattern, sp) 1919 out = bool(match) 1920 outs.append(out) 1921 # Print the matches for debugging 1922 print("pattern", pattern, "in:", strings_to_parse) 1923 print("matches", match, "result:", out) 1924 return outs 1925 1926def print_histogram( 1927 data, 1928 bins=10, 1929 height=10, 1930 logscale=False, 1931 minbin=0, 1932 horizontal=True, 1933 char="\U00002589", 1934 c=None, 1935 bold=True, 1936 title="histogram", 1937 spacer="", 1938) -> np.ndarray: 1939 """ 1940 Ascii histogram printing. 1941 1942 Input can be a `vedo.Volume` or `vedo.Mesh`. 1943 Returns the raw data before binning (useful when passing vtk objects). 1944 1945 Arguments: 1946 bins : (int) 1947 number of histogram bins 1948 height : (int) 1949 height of the histogram in character units 1950 logscale : (bool) 1951 use logscale for frequencies 1952 minbin : (int) 1953 ignore bins before minbin 1954 horizontal : (bool) 1955 show histogram horizontally 1956 char : (str) 1957 character to be used 1958 bold : (bool) 1959 use boldface 1960 title : (str) 1961 histogram title 1962 spacer : (str) 1963 horizontal spacer 1964 1965 Example: 1966 ```python 1967 from vedo import print_histogram 1968 import numpy as np 1969 d = np.random.normal(size=1000) 1970 data = print_histogram(d, c='b', logscale=True, title='my scalars') 1971 data = print_histogram(d, c='o') 1972 print(np.mean(data)) # data here is same as d 1973 ``` 1974  1975 """ 1976 # credits: http://pyinsci.blogspot.com/2009/10/ascii-histograms.html 1977 # adapted for vedo by M.Musy, 2019 1978 1979 if not horizontal: # better aspect ratio 1980 bins *= 2 1981 1982 try: 1983 data = vtk2numpy(data.dataset.GetPointData().GetScalars()) 1984 except AttributeError: 1985 # already an array 1986 data = np.asarray(data) 1987 1988 if isinstance(data, vtki.vtkImageData): 1989 dims = data.GetDimensions() 1990 nvx = min(100000, dims[0] * dims[1] * dims[2]) 1991 idxs = np.random.randint(0, min(dims), size=(nvx, 3)) 1992 data = [] 1993 for ix, iy, iz in idxs: 1994 d = data.GetScalarComponentAsFloat(ix, iy, iz, 0) 1995 data.append(d) 1996 data = np.array(data) 1997 1998 elif isinstance(data, vtki.vtkPolyData): 1999 arr = data.GetPointData().GetScalars() 2000 if not arr: 2001 arr = data.GetCellData().GetScalars() 2002 if not arr: 2003 return np.array([]) 2004 data = vtk2numpy(arr) 2005 2006 try: 2007 h = np.histogram(data, bins=bins) 2008 except TypeError as e: 2009 vedo.logger.error(f"cannot compute histogram: {e}") 2010 return np.array([]) 2011 2012 if minbin: 2013 hi = h[0][minbin:-1] 2014 else: 2015 hi = h[0] 2016 2017 if char == "\U00002589" and horizontal: 2018 char = "\U00002586" 2019 2020 title = title.ljust(14) + ":" 2021 entrs = " entries=" + str(len(data)) 2022 if logscale: 2023 h0 = np.log10(hi + 1) 2024 maxh0 = int(max(h0) * 100) / 100 2025 title = title + entrs + " (logscale)" 2026 else: 2027 h0 = hi 2028 maxh0 = max(h0) 2029 title = title + entrs 2030 2031 def _v(): 2032 his = "" 2033 if title: 2034 his += title + "\n" 2035 bars = h0 / maxh0 * height 2036 for l in reversed(range(1, height + 1)): 2037 line = "" 2038 if l == height: 2039 line = "%s " % maxh0 2040 else: 2041 line = " |" + " " * (len(str(maxh0)) - 3) 2042 for c in bars: 2043 if c >= np.ceil(l): 2044 line += char 2045 else: 2046 line += " " 2047 line += "\n" 2048 his += line 2049 his += "%.2f" % h[1][0] + "." * (bins) + "%.2f" % h[1][-1] + "\n" 2050 return his 2051 2052 def _h(): 2053 his = "" 2054 if title: 2055 his += title + "\n" 2056 xl = ["%.2f" % n for n in h[1]] 2057 lxl = [len(l) for l in xl] 2058 bars = h0 / maxh0 * height 2059 his += spacer + " " * int(max(bars) + 2 + max(lxl)) + "%s\n" % maxh0 2060 for i, c in enumerate(bars): 2061 line = xl[i] + " " * int(max(lxl) - lxl[i]) + "| " + char * int(c) + "\n" 2062 his += spacer + line 2063 return his 2064 2065 if horizontal: 2066 height *= 2 2067 vedo.printc(_h(), c=c, bold=bold) 2068 else: 2069 vedo.printc(_v(), c=c, bold=bold) 2070 return data 2071 2072 2073def print_table(*columns, headers=None, c="g") -> None: 2074 """ 2075 Print lists as tables. 2076 2077 Example: 2078 ```python 2079 from vedo.utils import print_table 2080 list1 = ["A", "B", "C"] 2081 list2 = [142, 220, 330] 2082 list3 = [True, False, True] 2083 headers = ["First Column", "Second Column", "Third Column"] 2084 print_table(list1, list2, list3, headers=headers) 2085 ``` 2086 2087  2088 """ 2089 # If headers is not provided, use default header names 2090 corner = "─" 2091 if headers is None: 2092 headers = [f"Column {i}" for i in range(1, len(columns) + 1)] 2093 assert len(headers) == len(columns) 2094 2095 # Find the maximum length of the elements in each column and header 2096 max_lens = [max(len(str(x)) for x in column) for column in columns] 2097 max_len_headers = [max(len(str(header)), max_len) for header, max_len in zip(headers, max_lens)] 2098 2099 # Construct the table header 2100 header = ( 2101 "│ " 2102 + " │ ".join(header.ljust(max_len) for header, max_len in zip(headers, max_len_headers)) 2103 + " │" 2104 ) 2105 2106 # Construct the line separator 2107 line1 = "┌" + corner.join("─" * (max_len + 2) for max_len in max_len_headers) + "┐" 2108 line2 = "└" + corner.join("─" * (max_len + 2) for max_len in max_len_headers) + "┘" 2109 2110 # Print the table header 2111 vedo.printc(line1, c=c) 2112 vedo.printc(header, c=c) 2113 vedo.printc(line2, c=c) 2114 2115 # Print the data rows 2116 for row in zip(*columns): 2117 row = ( 2118 "│ " 2119 + " │ ".join(str(col).ljust(max_len) for col, max_len in zip(row, max_len_headers)) 2120 + " │" 2121 ) 2122 vedo.printc(row, bold=False, c=c) 2123 2124 # Print the line separator again to close the table 2125 vedo.printc(line2, c=c) 2126 2127def print_inheritance_tree(C) -> None: 2128 """Prints the inheritance tree of class C.""" 2129 # Adapted from: https://stackoverflow.com/questions/26568976/ 2130 def class_tree(cls): 2131 subc = [class_tree(sub_class) for sub_class in cls.__subclasses__()] 2132 return {cls.__name__: subc} 2133 2134 def print_tree(tree, indent=8, current_ind=0): 2135 for k, v in tree.items(): 2136 if current_ind: 2137 before_dashes = current_ind - indent 2138 m = " " * before_dashes + "└" + "─" * (indent - 1) + " " + k 2139 vedo.printc(m) 2140 else: 2141 vedo.printc(k) 2142 for sub_tree in v: 2143 print_tree(sub_tree, indent=indent, current_ind=current_ind + indent) 2144 2145 if str(C.__class__) != "<class 'type'>": 2146 C = C.__class__ 2147 ct = class_tree(C) 2148 print_tree(ct) 2149 2150 2151def make_bands(inputlist, n): 2152 """ 2153 Group values of a list into bands of equal value, where 2154 `n` is the number of bands, a positive integer > 2. 2155 2156 Returns a binned list of the same length as the input. 2157 """ 2158 if n < 2: 2159 return inputlist 2160 vmin = np.min(inputlist) 2161 vmax = np.max(inputlist) 2162 bb = np.linspace(vmin, vmax, n, endpoint=0) 2163 dr = bb[1] - bb[0] 2164 bb += dr / 2 2165 tol = dr / 2 * 1.001 2166 newlist = [] 2167 for s in inputlist: 2168 for b in bb: 2169 if abs(s - b) < tol: 2170 newlist.append(b) 2171 break 2172 return np.array(newlist) 2173 2174 2175################################################################# 2176# Functions adapted from: 2177# https://github.com/sdorkenw/MeshParty/blob/master/meshparty/trimesh_vtk.py 2178def camera_from_quaternion(pos, quaternion, distance=10000, ngl_correct=True) -> vtki.vtkCamera: 2179 """ 2180 Define a `vtkCamera` with a particular orientation. 2181 2182 Arguments: 2183 pos: (np.array, list, tuple) 2184 an iterator of length 3 containing the focus point of the camera 2185 quaternion: (np.array, list, tuple) 2186 a len(4) quaternion `(x,y,z,w)` describing the rotation of the camera 2187 such as returned by neuroglancer `x,y,z,w` all in `[0,1]` range 2188 distance: (float) 2189 the desired distance from pos to the camera (default = 10000 nm) 2190 2191 Returns: 2192 `vtki.vtkCamera`, a vtk camera setup according to these rules. 2193 """ 2194 camera = vtki.vtkCamera() 2195 # define the quaternion in vtk, note the swapped order 2196 # w,x,y,z instead of x,y,z,w 2197 quat_vtk = vtki.get_class("Quaternion")( 2198 quaternion[3], quaternion[0], quaternion[1], quaternion[2]) 2199 # use this to define a rotation matrix in x,y,z 2200 # right handed units 2201 M = np.zeros((3, 3), dtype=np.float32) 2202 quat_vtk.ToMatrix3x3(M) 2203 # the default camera orientation is y up 2204 up = [0, 1, 0] 2205 # calculate default camera position is backed off in positive z 2206 pos = [0, 0, distance] 2207 2208 # set the camera rototation by applying the rotation matrix 2209 camera.SetViewUp(*np.dot(M, up)) 2210 # set the camera position by applying the rotation matrix 2211 camera.SetPosition(*np.dot(M, pos)) 2212 if ngl_correct: 2213 # neuroglancer has positive y going down 2214 # so apply these azimuth and roll corrections 2215 # to fix orientatins 2216 camera.Azimuth(-180) 2217 camera.Roll(180) 2218 2219 # shift the camera posiiton and focal position 2220 # to be centered on the desired location 2221 p = camera.GetPosition() 2222 p_new = np.array(p) + pos 2223 camera.SetPosition(*p_new) 2224 camera.SetFocalPoint(*pos) 2225 return camera 2226 2227 2228def camera_from_neuroglancer(state, zoom) -> vtki.vtkCamera: 2229 """ 2230 Define a `vtkCamera` from a neuroglancer state dictionary. 2231 2232 Arguments: 2233 state: (dict) 2234 an neuroglancer state dictionary. 2235 zoom: (float) 2236 how much to multiply zoom by to get camera backoff distance 2237 2238 Returns: 2239 `vtki.vtkCamera`, a vtk camera setup that matches this state. 2240 """ 2241 orient = state.get("perspectiveOrientation", [0.0, 0.0, 0.0, 1.0]) 2242 pzoom = state.get("perspectiveZoom", 10.0) 2243 position = state["navigation"]["pose"]["position"] 2244 pos_nm = np.array(position["voxelCoordinates"]) * position["voxelSize"] 2245 return camera_from_quaternion(pos_nm, orient, pzoom * zoom, ngl_correct=True) 2246 2247 2248def oriented_camera(center, up_vector, backoff_vector, backoff) -> vtki.vtkCamera: 2249 """ 2250 Generate a `vtkCamera` pointed at a specific location, 2251 oriented with a given up direction, set to a backoff. 2252 """ 2253 vup = np.array(up_vector) 2254 vup = vup / np.linalg.norm(vup) 2255 pt_backoff = center - backoff * np.array(backoff_vector) 2256 camera = vtki.vtkCamera() 2257 camera.SetFocalPoint(center[0], center[1], center[2]) 2258 camera.SetViewUp(vup[0], vup[1], vup[2]) 2259 camera.SetPosition(pt_backoff[0], pt_backoff[1], pt_backoff[2]) 2260 return camera 2261 2262 2263def camera_from_dict(camera, modify_inplace=None) -> vtki.vtkCamera: 2264 """ 2265 Generate a `vtkCamera` object from a python dictionary. 2266 2267 Parameters of the camera are: 2268 - `position` or `pos` (3-tuple) 2269 - `focal_point` (3-tuple) 2270 - `viewup` (3-tuple) 2271 - `distance` (float) 2272 - `clipping_range` (2-tuple) 2273 - `parallel_scale` (float) 2274 - `thickness` (float) 2275 - `view_angle` (float) 2276 - `roll` (float) 2277 2278 Exaplanation of the parameters can be found in the 2279 [vtkCamera documentation](https://vtk.org/doc/nightly/html/classvtkCamera.html). 2280 2281 Arguments: 2282 camera: (dict) 2283 a python dictionary containing camera parameters. 2284 modify_inplace: (vtkCamera) 2285 an existing `vtkCamera` object to modify in place. 2286 2287 Returns: 2288 `vtki.vtkCamera`, a vtk camera setup that matches this state. 2289 """ 2290 if modify_inplace: 2291 vcam = modify_inplace 2292 else: 2293 vcam = vtki.vtkCamera() 2294 2295 camera = dict(camera) # make a copy so input is not emptied by pop() 2296 2297 cm_pos = camera.pop("position", camera.pop("pos", None)) 2298 cm_focal_point = camera.pop("focal_point", camera.pop("focalPoint", None)) 2299 cm_viewup = camera.pop("viewup", None) 2300 cm_distance = camera.pop("distance", None) 2301 cm_clipping_range = camera.pop("clipping_range", camera.pop("clippingRange", None)) 2302 cm_parallel_scale = camera.pop("parallel_scale", camera.pop("parallelScale", None)) 2303 cm_thickness = camera.pop("thickness", None) 2304 cm_view_angle = camera.pop("view_angle", camera.pop("viewAngle", None)) 2305 cm_roll = camera.pop("roll", None) 2306 2307 if len(camera.keys()) > 0: 2308 vedo.logger.warning(f"in camera_from_dict, key(s) not recognized: {camera.keys()}") 2309 if cm_pos is not None: vcam.SetPosition(cm_pos) 2310 if cm_focal_point is not None: vcam.SetFocalPoint(cm_focal_point) 2311 if cm_viewup is not None: vcam.SetViewUp(cm_viewup) 2312 if cm_distance is not None: vcam.SetDistance(cm_distance) 2313 if cm_clipping_range is not None: vcam.SetClippingRange(cm_clipping_range) 2314 if cm_parallel_scale is not None: vcam.SetParallelScale(cm_parallel_scale) 2315 if cm_thickness is not None: vcam.SetThickness(cm_thickness) 2316 if cm_view_angle is not None: vcam.SetViewAngle(cm_view_angle) 2317 if cm_roll is not None: vcam.SetRoll(cm_roll) 2318 return vcam 2319 2320def camera_to_dict(vtkcam) -> dict: 2321 """ 2322 Convert a [vtkCamera](https://vtk.org/doc/nightly/html/classvtkCamera.html) 2323 object into a python dictionary. 2324 2325 Parameters of the camera are: 2326 - `position` (3-tuple) 2327 - `focal_point` (3-tuple) 2328 - `viewup` (3-tuple) 2329 - `distance` (float) 2330 - `clipping_range` (2-tuple) 2331 - `parallel_scale` (float) 2332 - `thickness` (float) 2333 - `view_angle` (float) 2334 - `roll` (float) 2335 2336 Arguments: 2337 vtkcam: (vtkCamera) 2338 a `vtkCamera` object to convert. 2339 """ 2340 cam = dict() 2341 cam["position"] = np.array(vtkcam.GetPosition()) 2342 cam["focal_point"] = np.array(vtkcam.GetFocalPoint()) 2343 cam["viewup"] = np.array(vtkcam.GetViewUp()) 2344 cam["distance"] = vtkcam.GetDistance() 2345 cam["clipping_range"] = np.array(vtkcam.GetClippingRange()) 2346 cam["parallel_scale"] = vtkcam.GetParallelScale() 2347 cam["thickness"] = vtkcam.GetThickness() 2348 cam["view_angle"] = vtkcam.GetViewAngle() 2349 cam["roll"] = vtkcam.GetRoll() 2350 return cam 2351 2352 2353def vtkCameraToK3D(vtkcam) -> np.ndarray: 2354 """ 2355 Convert a `vtkCamera` object into a 9-element list to be used by the K3D backend. 2356 2357 Output format is: 2358 `[posx,posy,posz, targetx,targety,targetz, upx,upy,upz]`. 2359 """ 2360 cpos = np.array(vtkcam.GetPosition()) 2361 kam = [cpos.tolist()] 2362 kam.append(vtkcam.GetFocalPoint()) 2363 kam.append(vtkcam.GetViewUp()) 2364 return np.array(kam).ravel() 2365 2366 2367def make_ticks( 2368 x0: float, 2369 x1: float, 2370 n=None, 2371 labels=None, 2372 digits=None, 2373 logscale=False, 2374 useformat="", 2375 ) -> Tuple[np.ndarray, List[str]]: 2376 """ 2377 Generate numeric labels for the `[x0, x1]` range. 2378 2379 The format specifier could be expressed in the format: 2380 `:[[fill]align][sign][#][0][width][,][.precision][type]` 2381 2382 where, the options are: 2383 ``` 2384 fill = any character 2385 align = < | > | = | ^ 2386 sign = + | - | " " 2387 width = integer 2388 precision = integer 2389 type = b | c | d | e | E | f | F | g | G | n | o | s | x | X | % 2390 ``` 2391 2392 E.g.: useformat=":.2f" 2393 """ 2394 # Copyright M. Musy, 2021, license: MIT. 2395 # 2396 # useformat eg: ":.2f", check out: 2397 # https://mkaz.blog/code/python-string-format-cookbook/ 2398 # https://www.programiz.com/python-programming/methods/built-in/format 2399 2400 if x1 <= x0: 2401 # vedo.printc("Error in make_ticks(): x0 >= x1", x0,x1, c='r') 2402 return np.array([0.0, 1.0]), ["", ""] 2403 2404 ticks_str, ticks_float = [], [] 2405 baseline = (1, 2, 5, 10, 20, 50) 2406 2407 if logscale: 2408 if x0 <= 0 or x1 <= 0: 2409 vedo.logger.error("make_ticks: zero or negative range with log scale.") 2410 raise RuntimeError 2411 if n is None: 2412 n = int(abs(np.log10(x1) - np.log10(x0))) + 1 2413 x0, x1 = np.log10([x0, x1]) 2414 2415 if not n: 2416 n = 5 2417 2418 if labels is not None: 2419 # user is passing custom labels 2420 2421 ticks_float.append(0) 2422 ticks_str.append("") 2423 for tp, ts in labels: 2424 if tp == x1: 2425 continue 2426 ticks_str.append(str(ts)) 2427 tickn = lin_interpolate(tp, [x0, x1], [0, 1]) 2428 ticks_float.append(tickn) 2429 2430 else: 2431 # ..here comes one of the shortest and most painful pieces of code: 2432 # automatically choose the best natural axis subdivision based on multiples of 1,2,5 2433 dstep = (x1 - x0) / n # desired step size, begin of the nightmare 2434 2435 basestep = pow(10, np.floor(np.log10(dstep))) 2436 steps = np.array([basestep * i for i in baseline]) 2437 idx = (np.abs(steps - dstep)).argmin() 2438 s = steps[idx] # chosen step size 2439 2440 low_bound, up_bound = 0, 0 2441 if x0 < 0: 2442 low_bound = -pow(10, np.ceil(np.log10(-x0))) 2443 if x1 > 0: 2444 up_bound = pow(10, np.ceil(np.log10(x1))) 2445 2446 if low_bound < 0: 2447 if up_bound < 0: 2448 negaxis = np.arange(low_bound, int(up_bound / s) * s) 2449 else: 2450 if -low_bound / s > 1.0e06: 2451 return np.array([0.0, 1.0]), ["", ""] 2452 negaxis = np.arange(low_bound, 0, s) 2453 else: 2454 negaxis = np.array([]) 2455 2456 if up_bound > 0: 2457 if low_bound > 0: 2458 posaxis = np.arange(int(low_bound / s) * s, up_bound, s) 2459 else: 2460 if up_bound / s > 1.0e06: 2461 return np.array([0.0, 1.0]), ["", ""] 2462 posaxis = np.arange(0, up_bound, s) 2463 else: 2464 posaxis = np.array([]) 2465 2466 fulaxis = np.unique(np.clip(np.concatenate([negaxis, posaxis]), x0, x1)) 2467 # end of the nightmare 2468 2469 if useformat: 2470 sf = "{" + f"{useformat}" + "}" 2471 sas = "" 2472 for x in fulaxis: 2473 sas += sf.format(x) + " " 2474 elif digits is None: 2475 np.set_printoptions(suppress=True) # avoid zero precision 2476 sas = str(fulaxis).replace("[", "").replace("]", "") 2477 sas = sas.replace(".e", "e").replace("e+0", "e+").replace("e-0", "e-") 2478 np.set_printoptions(suppress=None) # set back to default 2479 else: 2480 sas = precision(fulaxis, digits, vrange=(x0, x1)) 2481 sas = sas.replace("[", "").replace("]", "").replace(")", "").replace(",", "") 2482 2483 sas2 = [] 2484 for s in sas.split(): 2485 if s.endswith("."): 2486 s = s[:-1] 2487 if s == "-0": 2488 s = "0" 2489 if digits is not None and "e" in s: 2490 s += " " # add space to terminate modifiers 2491 sas2.append(s) 2492 2493 for ts, tp in zip(sas2, fulaxis): 2494 if tp == x1: 2495 continue 2496 tickn = lin_interpolate(tp, [x0, x1], [0, 1]) 2497 ticks_float.append(tickn) 2498 if logscale: 2499 val = np.power(10, tp) 2500 if useformat: 2501 sf = "{" + f"{useformat}" + "}" 2502 ticks_str.append(sf.format(val)) 2503 else: 2504 if val >= 10: 2505 val = int(val + 0.5) 2506 else: 2507 val = round_to_digit(val, 2) 2508 ticks_str.append(str(val)) 2509 else: 2510 ticks_str.append(ts) 2511 2512 ticks_str.append("") 2513 ticks_float.append(1) 2514 return np.array(ticks_float), ticks_str 2515 2516 2517def grid_corners(i: int, nm: list, size: list, margin=0, yflip=True) -> Tuple[np.ndarray, np.ndarray]: 2518 """ 2519 Compute the 2 corners coordinates of the i-th box in a grid of shape n*m. 2520 The top-left square is square number 1. 2521 2522 Arguments: 2523 i : (int) 2524 input index of the desired grid square (to be used in `show(..., at=...)`). 2525 nm : (list) 2526 grid shape as (n,m). 2527 size : (list) 2528 total size of the grid along x and y. 2529 margin : (float) 2530 keep a small margin between boxes. 2531 yflip : (bool) 2532 y-coordinate points downwards 2533 2534 Returns: 2535 Two 2D points representing the bottom-left corner and the top-right corner 2536 of the `i`-nth box in the grid. 2537 2538 Example: 2539 ```python 2540 from vedo import * 2541 acts=[] 2542 n,m = 5,7 2543 for i in range(1, n*m + 1): 2544 c1,c2 = utils.grid_corners(i, [n,m], [1,1], 0.01) 2545 t = Text3D(i, (c1+c2)/2, c='k', s=0.02, justify='center').z(0.01) 2546 r = Rectangle(c1, c2, c=i) 2547 acts += [t,r] 2548 show(acts, axes=1).close() 2549 ``` 2550  2551 """ 2552 i -= 1 2553 n, m = nm 2554 sx, sy = size 2555 dx, dy = sx / n, sy / m 2556 nx = i % n 2557 ny = int((i - nx) / n) 2558 if yflip: 2559 ny = n - ny 2560 c1 = (dx * nx + margin, dy * ny + margin) 2561 c2 = (dx * (nx + 1) - margin, dy * (ny + 1) - margin) 2562 return np.array(c1), np.array(c2) 2563 2564 2565############################################################################ 2566# Trimesh support 2567# 2568# Install trimesh with: 2569# 2570# sudo apt install python3-rtree 2571# pip install rtree shapely 2572# conda install trimesh 2573# 2574# Check the example gallery in: examples/other/trimesh> 2575########################################################################### 2576def vedo2trimesh(mesh): 2577 """ 2578 Convert `vedo.mesh.Mesh` to `Trimesh.Mesh` object. 2579 """ 2580 if is_sequence(mesh): 2581 tms = [] 2582 for a in mesh: 2583 tms.append(vedo2trimesh(a)) 2584 return tms 2585 2586 try: 2587 from trimesh import Trimesh # type: ignore 2588 except (ImportError, ModuleNotFoundError): 2589 vedo.logger.error("Need trimesh to run:\npip install trimesh") 2590 return None 2591 2592 tris = mesh.cells 2593 carr = mesh.celldata["CellIndividualColors"] 2594 ccols = carr 2595 2596 points = mesh.coordinates 2597 varr = mesh.pointdata["VertexColors"] 2598 vcols = varr 2599 2600 if len(tris) == 0: 2601 tris = None 2602 2603 return Trimesh(vertices=points, faces=tris, face_colors=ccols, vertex_colors=vcols, process=False) 2604 2605 2606def trimesh2vedo(inputobj): 2607 """ 2608 Convert a `Trimesh` object to `vedo.Mesh` or `vedo.Assembly` object. 2609 """ 2610 if is_sequence(inputobj): 2611 vms = [] 2612 for ob in inputobj: 2613 vms.append(trimesh2vedo(ob)) 2614 return vms 2615 2616 inputobj_type = str(type(inputobj)) 2617 2618 if "Trimesh" in inputobj_type or "primitives" in inputobj_type: 2619 faces = inputobj.faces 2620 poly = buildPolyData(inputobj.vertices, faces) 2621 tact = vedo.Mesh(poly) 2622 if inputobj.visual.kind == "face": 2623 trim_c = inputobj.visual.face_colors 2624 elif inputobj.visual.kind == "texture": 2625 trim_c = inputobj.visual.to_color().vertex_colors 2626 else: 2627 trim_c = inputobj.visual.vertex_colors 2628 2629 if is_sequence(trim_c): 2630 if is_sequence(trim_c[0]): 2631 same_color = len(np.unique(trim_c, axis=0)) < 2 # all vtxs have same color 2632 2633 if same_color: 2634 tact.c(trim_c[0, [0, 1, 2]]).alpha(trim_c[0, 3]) 2635 else: 2636 if inputobj.visual.kind == "face": 2637 tact.cellcolors = trim_c 2638 return tact 2639 2640 if "PointCloud" in inputobj_type: 2641 2642 vdpts = vedo.shapes.Points(inputobj.vertices, r=8, c='k') 2643 if hasattr(inputobj, "vertices_color"): 2644 vcols = (inputobj.vertices_color * 1).astype(np.uint8) 2645 vdpts.pointcolors = vcols 2646 return vdpts 2647 2648 if "path" in inputobj_type: 2649 2650 lines = [] 2651 for e in inputobj.entities: 2652 # print('trimesh entity', e.to_dict()) 2653 l = vedo.shapes.Line(inputobj.vertices[e.points], c="k", lw=2) 2654 lines.append(l) 2655 return vedo.Assembly(lines) 2656 2657 return None 2658 2659 2660def vedo2meshlab(vmesh): 2661 """Convert a `vedo.Mesh` to a Meshlab object.""" 2662 try: 2663 import pymeshlab as mlab # type: ignore 2664 except ModuleNotFoundError: 2665 vedo.logger.error("Need pymeshlab to run:\npip install pymeshlab") 2666 2667 vertex_matrix = vmesh.vertices.astype(np.float64) 2668 2669 try: 2670 face_matrix = np.asarray(vmesh.cells, dtype=np.float64) 2671 except: 2672 print("WARNING: in vedo2meshlab(), need to triangulate mesh first!") 2673 face_matrix = np.array(vmesh.clone().triangulate().cells, dtype=np.float64) 2674 2675 # v_normals_matrix = vmesh.normals(cells=False, recompute=False) 2676 v_normals_matrix = vmesh.vertex_normals 2677 if not v_normals_matrix.shape[0]: 2678 v_normals_matrix = np.empty((0, 3), dtype=np.float64) 2679 2680 # f_normals_matrix = vmesh.normals(cells=True, recompute=False) 2681 f_normals_matrix = vmesh.cell_normals 2682 if not f_normals_matrix.shape[0]: 2683 f_normals_matrix = np.empty((0, 3), dtype=np.float64) 2684 2685 v_color_matrix = vmesh.pointdata["RGBA"] 2686 if v_color_matrix is None: 2687 v_color_matrix = np.empty((0, 4), dtype=np.float64) 2688 else: 2689 v_color_matrix = v_color_matrix.astype(np.float64) / 255 2690 if v_color_matrix.shape[1] == 3: 2691 v_color_matrix = np.c_[ 2692 v_color_matrix, np.ones(v_color_matrix.shape[0], dtype=np.float64) 2693 ] 2694 2695 f_color_matrix = vmesh.celldata["RGBA"] 2696 if f_color_matrix is None: 2697 f_color_matrix = np.empty((0, 4), dtype=np.float64) 2698 else: 2699 f_color_matrix = f_color_matrix.astype(np.float64) / 255 2700 if f_color_matrix.shape[1] == 3: 2701 f_color_matrix = np.c_[ 2702 f_color_matrix, np.ones(f_color_matrix.shape[0], dtype=np.float64) 2703 ] 2704 2705 m = mlab.Mesh( 2706 vertex_matrix=vertex_matrix, 2707 face_matrix=face_matrix, 2708 v_normals_matrix=v_normals_matrix, 2709 f_normals_matrix=f_normals_matrix, 2710 v_color_matrix=v_color_matrix, 2711 f_color_matrix=f_color_matrix, 2712 ) 2713 2714 for k in vmesh.pointdata.keys(): 2715 data = vmesh.pointdata[k] 2716 if data is not None: 2717 if data.ndim == 1: # scalar 2718 m.add_vertex_custom_scalar_attribute(data.astype(np.float64), k) 2719 elif data.ndim == 2: # vectorial data 2720 if "tcoord" not in k.lower() and k not in ["Normals", "TextureCoordinates"]: 2721 m.add_vertex_custom_point_attribute(data.astype(np.float64), k) 2722 2723 for k in vmesh.celldata.keys(): 2724 data = vmesh.celldata[k] 2725 if data is not None: 2726 if data.ndim == 1: # scalar 2727 m.add_face_custom_scalar_attribute(data.astype(np.float64), k) 2728 elif data.ndim == 2 and k != "Normals": # vectorial data 2729 m.add_face_custom_point_attribute(data.astype(np.float64), k) 2730 2731 m.update_bounding_box() 2732 return m 2733 2734 2735def meshlab2vedo(mmesh, pointdata_keys=(), celldata_keys=()): 2736 """Convert a Meshlab object to `vedo.Mesh`.""" 2737 inputtype = str(type(mmesh)) 2738 2739 if "MeshSet" in inputtype: 2740 mmesh = mmesh.current_mesh() 2741 2742 mpoints, mcells = mmesh.vertex_matrix(), mmesh.face_matrix() 2743 if len(mcells) > 0: 2744 polydata = buildPolyData(mpoints, mcells) 2745 else: 2746 polydata = buildPolyData(mpoints, None) 2747 2748 if mmesh.has_vertex_scalar(): 2749 parr = mmesh.vertex_scalar_array() 2750 parr_vtk = numpy_to_vtk(parr) 2751 parr_vtk.SetName("MeshLabScalars") 2752 polydata.GetPointData().AddArray(parr_vtk) 2753 polydata.GetPointData().SetActiveScalars("MeshLabScalars") 2754 2755 if mmesh.has_face_scalar(): 2756 carr = mmesh.face_scalar_array() 2757 carr_vtk = numpy_to_vtk(carr) 2758 carr_vtk.SetName("MeshLabScalars") 2759 polydata.GetCellData().AddArray(carr_vtk) 2760 polydata.GetCellData().SetActiveScalars("MeshLabScalars") 2761 2762 for k in pointdata_keys: 2763 parr = mmesh.vertex_custom_scalar_attribute_array(k) 2764 parr_vtk = numpy_to_vtk(parr) 2765 parr_vtk.SetName(k) 2766 polydata.GetPointData().AddArray(parr_vtk) 2767 polydata.GetPointData().SetActiveScalars(k) 2768 2769 for k in celldata_keys: 2770 carr = mmesh.face_custom_scalar_attribute_array(k) 2771 carr_vtk = numpy_to_vtk(carr) 2772 carr_vtk.SetName(k) 2773 polydata.GetCellData().AddArray(carr_vtk) 2774 polydata.GetCellData().SetActiveScalars(k) 2775 2776 pnorms = mmesh.vertex_normal_matrix() 2777 if len(pnorms) > 0: 2778 polydata.GetPointData().SetNormals(numpy2vtk(pnorms, name="Normals")) 2779 2780 cnorms = mmesh.face_normal_matrix() 2781 if len(cnorms) > 0: 2782 polydata.GetCellData().SetNormals(numpy2vtk(cnorms, name="Normals")) 2783 return vedo.Mesh(polydata) 2784 2785 2786def open3d2vedo(o3d_mesh): 2787 """Convert `open3d.geometry.TriangleMesh` to a `vedo.Mesh`.""" 2788 m = vedo.Mesh([np.array(o3d_mesh.vertices), np.array(o3d_mesh.triangles)]) 2789 # TODO: could also check whether normals and color are present in 2790 # order to port with the above vertices/faces 2791 return m 2792 2793 2794def vedo2open3d(vedo_mesh): 2795 """ 2796 Return an `open3d.geometry.TriangleMesh` version of the current mesh. 2797 """ 2798 try: 2799 import open3d as o3d # type: ignore 2800 except RuntimeError: 2801 vedo.logger.error("Need open3d to run:\npip install open3d") 2802 2803 # create from numpy arrays 2804 o3d_mesh = o3d.geometry.TriangleMesh( 2805 vertices=o3d.utility.Vector3dVector(vedo_mesh.vertices), 2806 triangles=o3d.utility.Vector3iVector(vedo_mesh.cells), 2807 ) 2808 # TODO: need to add some if check here in case color and normals 2809 # info are not existing 2810 # o3d_mesh.vertex_colors = o3d.utility.Vector3dVector(vedo_mesh.pointdata["RGB"]/255) 2811 # o3d_mesh.vertex_normals= o3d.utility.Vector3dVector(vedo_mesh.pointdata["Normals"]) 2812 return o3d_mesh 2813 2814def vedo2madcad(vedo_mesh): 2815 """ 2816 Convert a `vedo.Mesh` to a `madcad.Mesh`. 2817 """ 2818 try: 2819 import madcad # type: ignore 2820 import numbers 2821 except ModuleNotFoundError: 2822 vedo.logger.error("Need madcad to run:\npip install pymadcad") 2823 2824 points = [madcad.vec3(*pt) for pt in vedo_mesh.vertices] 2825 faces = [madcad.vec3(*fc) for fc in vedo_mesh.cells] 2826 2827 options = {} 2828 for key, val in vedo_mesh.pointdata.items(): 2829 vec_type = f"vec{val.shape[-1]}" 2830 is_float = np.issubdtype(val.dtype, np.floating) 2831 madcad_dtype = getattr(madcad, f"f{vec_type}" if is_float else vec_type) 2832 options[key] = [madcad_dtype(v) for v in val] 2833 2834 madcad_mesh = madcad.Mesh(points=points, faces=faces, options=options) 2835 2836 return madcad_mesh 2837 2838 2839def madcad2vedo(madcad_mesh): 2840 """ 2841 Convert a `madcad.Mesh` to a `vedo.Mesh`. 2842 2843 A pointdata or celldata array named "tracks" is added to the output mesh, indicating 2844 the mesh region each point belongs to. 2845 2846 A metadata array named "madcad_groups" is added to the output mesh, indicating 2847 the mesh groups. 2848 2849 See [pymadcad website](https://pymadcad.readthedocs.io/en/latest/index.html) 2850 for more info. 2851 """ 2852 try: 2853 madcad_mesh = madcad_mesh["part"] 2854 except: 2855 pass 2856 2857 madp = [] 2858 for p in madcad_mesh.points: 2859 madp.append([float(p[0]), float(p[1]), float(p[2])]) 2860 madp = np.array(madp) 2861 2862 madf = [] 2863 try: 2864 for f in madcad_mesh.faces: 2865 madf.append([int(f[0]), int(f[1]), int(f[2])]) 2866 madf = np.array(madf).astype(np.uint16) 2867 except AttributeError: 2868 # print("no faces") 2869 pass 2870 2871 made = [] 2872 try: 2873 edges = madcad_mesh.edges 2874 for e in edges: 2875 made.append([int(e[0]), int(e[1])]) 2876 made = np.array(made).astype(np.uint16) 2877 except (AttributeError, TypeError): 2878 # print("no edges") 2879 pass 2880 2881 try: 2882 line = np.array(madcad_mesh.indices).astype(np.uint16) 2883 made.append(line) 2884 except AttributeError: 2885 # print("no indices") 2886 pass 2887 2888 madt = [] 2889 try: 2890 for t in madcad_mesh.tracks: 2891 madt.append(int(t)) 2892 madt = np.array(madt).astype(np.uint16) 2893 except AttributeError: 2894 # print("no tracks") 2895 pass 2896 2897 ############################### 2898 poly = vedo.utils.buildPolyData(madp, madf, made) 2899 if len(madf) == 0 and len(made) == 0: 2900 m = vedo.Points(poly) 2901 else: 2902 m = vedo.Mesh(poly) 2903 2904 if len(madt) == len(madf): 2905 m.celldata["tracks"] = madt 2906 maxt = np.max(madt) 2907 m.mapper.SetScalarRange(0, np.max(madt)) 2908 if maxt==0: m.mapper.SetScalarVisibility(0) 2909 elif len(madt) == len(madp): 2910 m.pointdata["tracks"] = madt 2911 maxt = np.max(madt) 2912 m.mapper.SetScalarRange(0, maxt) 2913 if maxt==0: m.mapper.SetScalarVisibility(0) 2914 2915 try: 2916 m.info["madcad_groups"] = madcad_mesh.groups 2917 except AttributeError: 2918 # print("no groups") 2919 pass 2920 2921 try: 2922 options = dict(madcad_mesh.options) 2923 if "display_wire" in options and options["display_wire"]: 2924 m.lw(1).lc(madcad_mesh.c()) 2925 if "display_faces" in options and not options["display_faces"]: 2926 m.alpha(0.2) 2927 if "color" in options: 2928 m.c(options["color"]) 2929 2930 for key, val in options.items(): 2931 m.pointdata[key] = val 2932 2933 except AttributeError: 2934 # print("no options") 2935 pass 2936 2937 return m 2938 2939 2940def vtk_version_at_least(major, minor=0, build=0) -> bool: 2941 """ 2942 Check the installed VTK version. 2943 2944 Return `True` if the requested VTK version is greater or equal to the actual VTK version. 2945 2946 Arguments: 2947 major : (int) 2948 Major version. 2949 minor : (int) 2950 Minor version. 2951 build : (int) 2952 Build version. 2953 """ 2954 needed_version = 10000000000 * int(major) + 100000000 * int(minor) + int(build) 2955 try: 2956 vtk_version_number = vtki.VTK_VERSION_NUMBER 2957 except AttributeError: # as error: 2958 ver = vtki.vtkVersion() 2959 vtk_version_number = ( 2960 10000000000 * ver.GetVTKMajorVersion() 2961 + 100000000 * ver.GetVTKMinorVersion() 2962 + ver.GetVTKBuildVersion() 2963 ) 2964 return vtk_version_number >= needed_version 2965 2966 2967def ctf2lut(vol, logscale=False): 2968 """Internal use.""" 2969 # build LUT from a color transfer function for tmesh or volume 2970 2971 ctf = vol.properties.GetRGBTransferFunction() 2972 otf = vol.properties.GetScalarOpacity() 2973 x0, x1 = vol.dataset.GetScalarRange() 2974 cols, alphas = [], [] 2975 for x in np.linspace(x0, x1, 256): 2976 cols.append(ctf.GetColor(x)) 2977 alphas.append(otf.GetValue(x)) 2978 2979 if logscale: 2980 lut = vtki.vtkLogLookupTable() 2981 else: 2982 lut = vtki.vtkLookupTable() 2983 2984 lut.SetRange(x0, x1) 2985 lut.SetNumberOfTableValues(len(cols)) 2986 for i, col in enumerate(cols): 2987 r, g, b = col 2988 lut.SetTableValue(i, r, g, b, alphas[i]) 2989 lut.Build() 2990 return lut 2991 2992 2993def get_vtk_name_event(name: str) -> str: 2994 """ 2995 Return the name of a VTK event. 2996 2997 Frequently used events are: 2998 - KeyPress, KeyRelease: listen to keyboard events 2999 - LeftButtonPress, LeftButtonRelease: listen to mouse clicks 3000 - MiddleButtonPress, MiddleButtonRelease 3001 - RightButtonPress, RightButtonRelease 3002 - MouseMove: listen to mouse pointer changing position 3003 - MouseWheelForward, MouseWheelBackward 3004 - Enter, Leave: listen to mouse entering or leaving the window 3005 - Pick, StartPick, EndPick: listen to object picking 3006 - ResetCamera, ResetCameraClippingRange 3007 - Error, Warning, Char, Timer 3008 3009 Check the complete list of events here: 3010 https://vtk.org/doc/nightly/html/classvtkCommand.html 3011 """ 3012 # as vtk names are ugly and difficult to remember: 3013 ln = name.lower() 3014 if "click" in ln or "button" in ln: 3015 event_name = "LeftButtonPress" 3016 if "right" in ln: 3017 event_name = "RightButtonPress" 3018 elif "mid" in ln: 3019 event_name = "MiddleButtonPress" 3020 if "release" in ln: 3021 event_name = event_name.replace("Press", "Release") 3022 else: 3023 event_name = name 3024 if "key" in ln: 3025 if "release" in ln: 3026 event_name = "KeyRelease" 3027 else: 3028 event_name = "KeyPress" 3029 3030 if ("mouse" in ln and "mov" in ln) or "over" in ln: 3031 event_name = "MouseMove" 3032 3033 words = [ 3034 "pick", "timer", "reset", "enter", "leave", "char", 3035 "error", "warning", "start", "end", "wheel", "clipping", 3036 "range", "camera", "render", "interaction", "modified", 3037 ] 3038 for w in words: 3039 if w in ln: 3040 event_name = event_name.replace(w, w.capitalize()) 3041 3042 event_name = event_name.replace("REnd ", "Rend") 3043 event_name = event_name.replace("the ", "") 3044 event_name = event_name.replace(" of ", "").replace(" ", "") 3045 3046 if not event_name.endswith("Event"): 3047 event_name += "Event" 3048 3049 if vtki.vtkCommand.GetEventIdFromString(event_name) == 0: 3050 vedo.printc( 3051 f"Error: '{name}' is not a valid event name.", c='r') 3052 vedo.printc("Check the list of events here:", c='r') 3053 vedo.printc("\thttps://vtk.org/doc/nightly/html/classvtkCommand.html", c='r') 3054 # raise RuntimeError 3055 3056 return event_name
62class OperationNode: 63 """ 64 Keep track of the operations which led to a final state. 65 """ 66 # https://www.graphviz.org/doc/info/shapes.html#html 67 # Mesh #e9c46a 68 # Follower #d9ed92 69 # Volume, UnstructuredGrid #4cc9f0 70 # TetMesh #9e2a2b 71 # File #8a817c 72 # Image #f28482 73 # Assembly #f08080 74 75 def __init__( 76 self, operation, parents=(), comment="", shape="none", c="#e9c46a", style="filled" 77 ) -> None: 78 """ 79 Keep track of the operations which led to a final object. 80 This allows to show the `pipeline` tree for any `vedo` object with e.g.: 81 82 ```python 83 from vedo import * 84 sp = Sphere() 85 sp.clean().subdivide() 86 sp.pipeline.show() 87 ``` 88 89 Arguments: 90 operation : (str, class) 91 descriptor label, if a class is passed then grab its name 92 parents : (list) 93 list of the parent classes the object comes from 94 comment : (str) 95 a second-line text description 96 shape : (str) 97 shape of the frame, check out [this link.](https://graphviz.org/doc/info/shapes.html) 98 c : (hex) 99 hex color 100 style : (str) 101 comma-separated list of styles 102 103 Example: 104 ```python 105 from vedo.utils import OperationNode 106 107 op_node1 = OperationNode("Operation1", c="lightblue") 108 op_node2 = OperationNode("Operation2") 109 op_node3 = OperationNode("Operation3", shape='diamond') 110 op_node4 = OperationNode("Operation4") 111 op_node5 = OperationNode("Operation5") 112 op_node6 = OperationNode("Result", c="lightgreen") 113 114 op_node3.add_parent(op_node1) 115 op_node4.add_parent(op_node1) 116 op_node3.add_parent(op_node2) 117 op_node5.add_parent(op_node2) 118 op_node6.add_parent(op_node3) 119 op_node6.add_parent(op_node5) 120 op_node6.add_parent(op_node1) 121 122 op_node6.show(orientation="TB") 123 ``` 124  125 """ 126 if not vedo.settings.enable_pipeline: 127 return 128 129 if isinstance(operation, str): 130 self.operation = operation 131 else: 132 self.operation = operation.__class__.__name__ 133 self.operation_plain = str(self.operation) 134 135 pp = [] # filter out invalid stuff 136 for p in parents: 137 if hasattr(p, "pipeline"): 138 pp.append(p.pipeline) 139 self.parents = pp 140 141 if comment: 142 self.operation = f"<{self.operation}<BR/><SUB><I>{comment}</I></SUB>>" 143 144 self.dot = None 145 self.time = time.time() 146 self.shape = shape 147 self.style = style 148 self.color = c 149 self.counts = 0 150 151 def add_parent(self, parent) -> None: 152 """Add a parent to the list.""" 153 self.parents.append(parent) 154 155 def _build_tree(self, dot): 156 dot.node( 157 str(id(self)), 158 label=self.operation, 159 shape=self.shape, 160 color=self.color, 161 style=self.style, 162 ) 163 for parent in self.parents: 164 if parent: 165 t = f"{self.time - parent.time: .1f}s" 166 dot.edge(str(id(parent)), str(id(self)), label=t) 167 parent._build_tree(dot) 168 169 def __repr__(self): 170 try: 171 from treelib import Tree 172 except ImportError: 173 vedo.logger.error( 174 "To use this functionality please install treelib:" 175 "\n pip install treelib" 176 ) 177 return "" 178 179 def _build_tree(parent): 180 for par in parent.parents: 181 if par: 182 op = par.operation_plain 183 tree.create_node( 184 op, op + str(par.time), parent=parent.operation_plain + str(parent.time) 185 ) 186 _build_tree(par) 187 try: 188 tree = Tree() 189 tree.create_node(self.operation_plain, self.operation_plain + str(self.time)) 190 _build_tree(self) 191 out = tree.show(stdout=False) 192 except: 193 out = f"Sorry treelib failed to build the tree for '{self.operation_plain}()'." 194 return out 195 196 def print(self) -> None: 197 """Print the tree of operations.""" 198 print(self.__str__()) 199 200 def show(self, orientation="LR", popup=True) -> None: 201 """Show the graphviz output for the pipeline of this object""" 202 if not vedo.settings.enable_pipeline: 203 return 204 205 try: 206 from graphviz import Digraph 207 except ImportError: 208 vedo.logger.error("please install graphviz with command\n pip install graphviz") 209 vedo.logger.error(" sudo apt-get install graphviz -y") 210 return 211 212 # visualize the entire tree 213 dot = Digraph( 214 node_attr={"fontcolor": "#201010", "fontname": "Helvetica", "fontsize": "12"}, 215 edge_attr={"fontname": "Helvetica", "fontsize": "6", "arrowsize": "0.4"}, 216 ) 217 dot.attr(rankdir=orientation) 218 219 self.counts = 0 220 self._build_tree(dot) 221 self.dot = dot 222 223 home_dir = os.path.expanduser("~") 224 gpath = os.path.join( 225 home_dir, vedo.settings.cache_directory, "vedo", "pipeline_graphviz") 226 227 dot.render(gpath, view=popup)
Keep track of the operations which led to a final state.
75 def __init__( 76 self, operation, parents=(), comment="", shape="none", c="#e9c46a", style="filled" 77 ) -> None: 78 """ 79 Keep track of the operations which led to a final object. 80 This allows to show the `pipeline` tree for any `vedo` object with e.g.: 81 82 ```python 83 from vedo import * 84 sp = Sphere() 85 sp.clean().subdivide() 86 sp.pipeline.show() 87 ``` 88 89 Arguments: 90 operation : (str, class) 91 descriptor label, if a class is passed then grab its name 92 parents : (list) 93 list of the parent classes the object comes from 94 comment : (str) 95 a second-line text description 96 shape : (str) 97 shape of the frame, check out [this link.](https://graphviz.org/doc/info/shapes.html) 98 c : (hex) 99 hex color 100 style : (str) 101 comma-separated list of styles 102 103 Example: 104 ```python 105 from vedo.utils import OperationNode 106 107 op_node1 = OperationNode("Operation1", c="lightblue") 108 op_node2 = OperationNode("Operation2") 109 op_node3 = OperationNode("Operation3", shape='diamond') 110 op_node4 = OperationNode("Operation4") 111 op_node5 = OperationNode("Operation5") 112 op_node6 = OperationNode("Result", c="lightgreen") 113 114 op_node3.add_parent(op_node1) 115 op_node4.add_parent(op_node1) 116 op_node3.add_parent(op_node2) 117 op_node5.add_parent(op_node2) 118 op_node6.add_parent(op_node3) 119 op_node6.add_parent(op_node5) 120 op_node6.add_parent(op_node1) 121 122 op_node6.show(orientation="TB") 123 ``` 124  125 """ 126 if not vedo.settings.enable_pipeline: 127 return 128 129 if isinstance(operation, str): 130 self.operation = operation 131 else: 132 self.operation = operation.__class__.__name__ 133 self.operation_plain = str(self.operation) 134 135 pp = [] # filter out invalid stuff 136 for p in parents: 137 if hasattr(p, "pipeline"): 138 pp.append(p.pipeline) 139 self.parents = pp 140 141 if comment: 142 self.operation = f"<{self.operation}<BR/><SUB><I>{comment}</I></SUB>>" 143 144 self.dot = None 145 self.time = time.time() 146 self.shape = shape 147 self.style = style 148 self.color = c 149 self.counts = 0
Keep track of the operations which led to a final object.
This allows to show the pipeline tree for any vedo object with e.g.:
from vedo import *
sp = Sphere()
sp.clean().subdivide()
sp.pipeline.show()
Arguments:
- operation : (str, class) descriptor label, if a class is passed then grab its name
- parents : (list) list of the parent classes the object comes from
- comment : (str) a second-line text description
- shape : (str) shape of the frame, check out this link.
- c : (hex) hex color
- style : (str) comma-separated list of styles
Example:
from vedo.utils import OperationNode op_node1 = OperationNode("Operation1", c="lightblue") op_node2 = OperationNode("Operation2") op_node3 = OperationNode("Operation3", shape='diamond') op_node4 = OperationNode("Operation4") op_node5 = OperationNode("Operation5") op_node6 = OperationNode("Result", c="lightgreen") op_node3.add_parent(op_node1) op_node4.add_parent(op_node1) op_node3.add_parent(op_node2) op_node5.add_parent(op_node2) op_node6.add_parent(op_node3) op_node6.add_parent(op_node5) op_node6.add_parent(op_node1) op_node6.show(orientation="TB")
151 def add_parent(self, parent) -> None: 152 """Add a parent to the list.""" 153 self.parents.append(parent)
Add a parent to the list.
200 def show(self, orientation="LR", popup=True) -> None: 201 """Show the graphviz output for the pipeline of this object""" 202 if not vedo.settings.enable_pipeline: 203 return 204 205 try: 206 from graphviz import Digraph 207 except ImportError: 208 vedo.logger.error("please install graphviz with command\n pip install graphviz") 209 vedo.logger.error(" sudo apt-get install graphviz -y") 210 return 211 212 # visualize the entire tree 213 dot = Digraph( 214 node_attr={"fontcolor": "#201010", "fontname": "Helvetica", "fontsize": "12"}, 215 edge_attr={"fontname": "Helvetica", "fontsize": "6", "arrowsize": "0.4"}, 216 ) 217 dot.attr(rankdir=orientation) 218 219 self.counts = 0 220 self._build_tree(dot) 221 self.dot = dot 222 223 home_dir = os.path.expanduser("~") 224 gpath = os.path.join( 225 home_dir, vedo.settings.cache_directory, "vedo", "pipeline_graphviz") 226 227 dot.render(gpath, view=popup)
Show the graphviz output for the pipeline of this object
231class ProgressBar: 232 """ 233 Class to print a progress bar. 234 """ 235 236 def __init__( 237 self, 238 start, 239 stop, 240 step=1, 241 c=None, 242 bold=True, 243 italic=False, 244 title="", 245 eta=True, 246 delay=-1, 247 width=25, 248 char="\U00002501", 249 char_back="\U00002500", 250 ) -> None: 251 """ 252 Class to print a progress bar with optional text message. 253 254 Check out also function `progressbar()`. 255 256 Arguments: 257 start : (int) 258 starting value 259 stop : (int) 260 stopping value 261 step : (int) 262 step value 263 c : (str) 264 color in hex format 265 title : (str) 266 title text 267 eta : (bool) 268 estimate time of arrival 269 delay : (float) 270 minimum time before printing anything, 271 if negative use the default value 272 as set in `vedo.settings.progressbar_delay` 273 width : (int) 274 width of the progress bar 275 char : (str) 276 character to use for the progress bar 277 char_back : (str) 278 character to use for the background of the progress bar 279 280 Example: 281 ```python 282 import time 283 from vedo import ProgressBar 284 pb = ProgressBar(0,40, c='r') 285 for i in pb.range(): 286 time.sleep(0.1) 287 pb.print() 288 ``` 289  290 """ 291 self.char = char 292 self.char_back = char_back 293 294 self.title = title + " " 295 if title: 296 self.title = " " + self.title 297 298 if delay < 0: 299 delay = vedo.settings.progressbar_delay 300 301 self.start = start 302 self.stop = stop 303 self.step = step 304 305 self.color = c 306 self.bold = bold 307 self.italic = italic 308 self.width = width 309 self.pbar = "" 310 self.percent = 0.0 311 self.percent_int = 0 312 self.eta = eta 313 self.delay = delay 314 315 self.t0 = time.time() 316 self._remaining = 1e10 317 318 self._update(0) 319 320 self._counts = 0 321 self._oldbar = "" 322 self._lentxt = 0 323 self._range = np.arange(start, stop, step) 324 325 def print(self, txt="", c=None) -> None: 326 """Print the progress bar with an optional message.""" 327 if not c: 328 c = self.color 329 330 self._update(self._counts + self.step) 331 332 if self.delay: 333 if time.time() - self.t0 < self.delay: 334 return 335 336 if self.pbar != self._oldbar: 337 self._oldbar = self.pbar 338 339 if self.eta and self._counts > 1: 340 341 tdenom = time.time() - self.t0 342 if tdenom: 343 vel = self._counts / tdenom 344 self._remaining = (self.stop - self._counts) / vel 345 else: 346 vel = 1 347 self._remaining = 0.0 348 349 if self._remaining > 60: 350 _mins = int(self._remaining / 60) 351 _secs = self._remaining - 60 * _mins 352 mins = f"{_mins}m" 353 secs = f"{int(_secs + 0.5)}s " 354 else: 355 mins = "" 356 secs = f"{int(self._remaining + 0.5)}s " 357 358 vel = round(vel, 1) 359 eta = f"eta: {mins}{secs}({vel} it/s) " 360 if self._remaining < 0.5: 361 dt = time.time() - self.t0 362 if dt > 60: 363 _mins = int(dt / 60) 364 _secs = dt - 60 * _mins 365 mins = f"{_mins}m" 366 secs = f"{int(_secs + 0.5)}s " 367 else: 368 mins = "" 369 secs = f"{int(dt + 0.5)}s " 370 eta = f"elapsed: {mins}{secs}({vel} it/s) " 371 txt = "" 372 else: 373 eta = "" 374 375 eraser = " " * self._lentxt + "\b" * self._lentxt 376 377 s = f"{self.pbar} {eraser}{eta}{txt}\r" 378 vedo.printc(s, c=c, bold=self.bold, italic=self.italic, end="") 379 if self.percent > 99.999: 380 print("") 381 382 self._lentxt = len(txt) 383 384 def range(self) -> np.ndarray: 385 """Return the range iterator.""" 386 return self._range 387 388 def _update(self, counts): 389 if counts < self.start: 390 counts = self.start 391 elif counts > self.stop: 392 counts = self.stop 393 self._counts = counts 394 395 self.percent = (self._counts - self.start) * 100.0 396 397 delta = self.stop - self.start 398 if delta: 399 self.percent /= delta 400 else: 401 self.percent = 0.0 402 403 self.percent_int = int(round(self.percent)) 404 af = self.width - 2 405 nh = int(round(self.percent_int / 100 * af)) 406 pbar_background = "\x1b[2m" + self.char_back * (af - nh) 407 self.pbar = f"{self.title}{self.char * (nh-1)}{pbar_background}" 408 if self.percent < 100.0: 409 ps = f" {self.percent_int}%" 410 else: 411 ps = "" 412 self.pbar += ps
Class to print a progress bar.
236 def __init__( 237 self, 238 start, 239 stop, 240 step=1, 241 c=None, 242 bold=True, 243 italic=False, 244 title="", 245 eta=True, 246 delay=-1, 247 width=25, 248 char="\U00002501", 249 char_back="\U00002500", 250 ) -> None: 251 """ 252 Class to print a progress bar with optional text message. 253 254 Check out also function `progressbar()`. 255 256 Arguments: 257 start : (int) 258 starting value 259 stop : (int) 260 stopping value 261 step : (int) 262 step value 263 c : (str) 264 color in hex format 265 title : (str) 266 title text 267 eta : (bool) 268 estimate time of arrival 269 delay : (float) 270 minimum time before printing anything, 271 if negative use the default value 272 as set in `vedo.settings.progressbar_delay` 273 width : (int) 274 width of the progress bar 275 char : (str) 276 character to use for the progress bar 277 char_back : (str) 278 character to use for the background of the progress bar 279 280 Example: 281 ```python 282 import time 283 from vedo import ProgressBar 284 pb = ProgressBar(0,40, c='r') 285 for i in pb.range(): 286 time.sleep(0.1) 287 pb.print() 288 ``` 289  290 """ 291 self.char = char 292 self.char_back = char_back 293 294 self.title = title + " " 295 if title: 296 self.title = " " + self.title 297 298 if delay < 0: 299 delay = vedo.settings.progressbar_delay 300 301 self.start = start 302 self.stop = stop 303 self.step = step 304 305 self.color = c 306 self.bold = bold 307 self.italic = italic 308 self.width = width 309 self.pbar = "" 310 self.percent = 0.0 311 self.percent_int = 0 312 self.eta = eta 313 self.delay = delay 314 315 self.t0 = time.time() 316 self._remaining = 1e10 317 318 self._update(0) 319 320 self._counts = 0 321 self._oldbar = "" 322 self._lentxt = 0 323 self._range = np.arange(start, stop, step)
Class to print a progress bar with optional text message.
Check out also function progressbar().
Arguments:
- start : (int) starting value
- stop : (int) stopping value
- step : (int) step value
- c : (str) color in hex format
- title : (str) title text
- eta : (bool) estimate time of arrival
- delay : (float)
minimum time before printing anything,
if negative use the default value
as set in
vedo.settings.progressbar_delay - width : (int) width of the progress bar
- char : (str) character to use for the progress bar
- char_back : (str) character to use for the background of the progress bar
Example:
import time from vedo import ProgressBar pb = ProgressBar(0,40, c='r') for i in pb.range(): time.sleep(0.1) pb.print()
325 def print(self, txt="", c=None) -> None: 326 """Print the progress bar with an optional message.""" 327 if not c: 328 c = self.color 329 330 self._update(self._counts + self.step) 331 332 if self.delay: 333 if time.time() - self.t0 < self.delay: 334 return 335 336 if self.pbar != self._oldbar: 337 self._oldbar = self.pbar 338 339 if self.eta and self._counts > 1: 340 341 tdenom = time.time() - self.t0 342 if tdenom: 343 vel = self._counts / tdenom 344 self._remaining = (self.stop - self._counts) / vel 345 else: 346 vel = 1 347 self._remaining = 0.0 348 349 if self._remaining > 60: 350 _mins = int(self._remaining / 60) 351 _secs = self._remaining - 60 * _mins 352 mins = f"{_mins}m" 353 secs = f"{int(_secs + 0.5)}s " 354 else: 355 mins = "" 356 secs = f"{int(self._remaining + 0.5)}s " 357 358 vel = round(vel, 1) 359 eta = f"eta: {mins}{secs}({vel} it/s) " 360 if self._remaining < 0.5: 361 dt = time.time() - self.t0 362 if dt > 60: 363 _mins = int(dt / 60) 364 _secs = dt - 60 * _mins 365 mins = f"{_mins}m" 366 secs = f"{int(_secs + 0.5)}s " 367 else: 368 mins = "" 369 secs = f"{int(dt + 0.5)}s " 370 eta = f"elapsed: {mins}{secs}({vel} it/s) " 371 txt = "" 372 else: 373 eta = "" 374 375 eraser = " " * self._lentxt + "\b" * self._lentxt 376 377 s = f"{self.pbar} {eraser}{eta}{txt}\r" 378 vedo.printc(s, c=c, bold=self.bold, italic=self.italic, end="") 379 if self.percent > 99.999: 380 print("") 381 382 self._lentxt = len(txt)
Print the progress bar with an optional message.
416def progressbar( 417 iterable, 418 c=None, bold=True, italic=False, title="", 419 eta=True, width=25, delay=-1, 420 ): 421 """ 422 Function to print a progress bar with optional text message. 423 424 Use delay to set a minimum time before printing anything. 425 If delay is negative, then use the default value 426 as set in `vedo.settings.progressbar_delay`. 427 428 Arguments: 429 start : (int) 430 starting value 431 stop : (int) 432 stopping value 433 step : (int) 434 step value 435 c : (str) 436 color in hex format 437 title : (str) 438 title text 439 eta : (bool) 440 estimate time of arrival 441 delay : (float) 442 minimum time before printing anything, 443 if negative use the default value 444 set in `vedo.settings.progressbar_delay` 445 width : (int) 446 width of the progress bar 447 char : (str) 448 character to use for the progress bar 449 char_back : (str) 450 character to use for the background of the progress bar 451 452 Example: 453 ```python 454 import time 455 for i in progressbar(range(100), c='r'): 456 time.sleep(0.1) 457 ``` 458  459 """ 460 try: 461 if is_number(iterable): 462 total = int(iterable) 463 iterable = range(total) 464 else: 465 total = len(iterable) 466 except TypeError: 467 iterable = list(iterable) 468 total = len(iterable) 469 470 pb = ProgressBar( 471 0, total, c=c, bold=bold, italic=italic, title=title, 472 eta=eta, delay=delay, width=width, 473 ) 474 for item in iterable: 475 pb.print() 476 yield item
Function to print a progress bar with optional text message.
Use delay to set a minimum time before printing anything.
If delay is negative, then use the default value
as set in vedo.settings.progressbar_delay.
Arguments:
- start : (int) starting value
- stop : (int) stopping value
- step : (int) step value
- c : (str) color in hex format
- title : (str) title text
- eta : (bool) estimate time of arrival
- delay : (float)
minimum time before printing anything,
if negative use the default value
set in
vedo.settings.progressbar_delay - width : (int) width of the progress bar
- char : (str) character to use for the progress bar
- char_back : (str) character to use for the background of the progress bar
Example:
import time for i in progressbar(range(100), c='r'): time.sleep(0.1)
480class Minimizer: 481 """ 482 A function minimizer that uses the Nelder-Mead method. 483 484 The algorithm constructs an n-dimensional simplex in parameter 485 space (i.e. a tetrahedron if the number or parameters is 3) 486 and moves the vertices around parameter space until 487 a local minimum is found. The amoeba method is robust, 488 reasonably efficient, but is not guaranteed to find 489 the global minimum if several local minima exist. 490 491 Arguments: 492 function : (callable) 493 the function to minimize 494 max_iterations : (int) 495 the maximum number of iterations 496 contraction_ratio : (float) 497 The contraction ratio. 498 The default value of 0.5 gives fast convergence, 499 but larger values such as 0.6 or 0.7 provide greater stability. 500 expansion_ratio : (float) 501 The expansion ratio. 502 The default value is 2.0, which provides rapid expansion. 503 Values between 1.1 and 2.0 are valid. 504 tol : (float) 505 the tolerance for convergence 506 507 Example: 508 - [nelder-mead.py](https://github.com/marcomusy/vedo/blob/master/examples/others/nelder-mead.py) 509 """ 510 def __init__( 511 self, 512 function=None, 513 max_iterations=10000, 514 contraction_ratio=0.5, 515 expansion_ratio=2.0, 516 tol=1e-5, 517 ) -> None: 518 self.function = function 519 self.tolerance = tol 520 self.contraction_ratio = contraction_ratio 521 self.expansion_ratio = expansion_ratio 522 self.max_iterations = max_iterations 523 self.minimizer = vtki.new("AmoebaMinimizer") 524 self.minimizer.SetFunction(self._vtkfunc) 525 self.results = {} 526 self.parameters_path = [] 527 self.function_path = [] 528 529 def _vtkfunc(self): 530 n = self.minimizer.GetNumberOfParameters() 531 ain = [self.minimizer.GetParameterValue(i) for i in range(n)] 532 r = self.function(ain) 533 self.minimizer.SetFunctionValue(r) 534 self.parameters_path.append(ain) 535 self.function_path.append(r) 536 return r 537 538 def eval(self, parameters=()) -> float: 539 """ 540 Evaluate the function at the current or given parameters. 541 """ 542 if len(parameters) == 0: 543 return self.minimizer.EvaluateFunction() 544 self.set_parameters(parameters) 545 return self.function(parameters) 546 547 def set_parameter(self, name, value, scale=1.0) -> None: 548 """ 549 Set the parameter value. 550 The initial amount by which the parameter 551 will be modified during the search for the minimum. 552 """ 553 self.minimizer.SetParameterValue(name, value) 554 self.minimizer.SetParameterScale(name, scale) 555 556 def set_parameters(self, parameters) -> None: 557 """ 558 Set the parameters names and values from a dictionary. 559 """ 560 for name, value in parameters.items(): 561 if len(value) == 2: 562 self.set_parameter(name, value[0], value[1]) 563 else: 564 self.set_parameter(name, value) 565 566 def minimize(self) -> dict: 567 """ 568 Minimize the input function. 569 570 A dictionary with the minimization results 571 including the initial parameters, final parameters, 572 minimum value, number of iterations, maximum iterations, 573 tolerance, convergence flag, parameters path, 574 function path, Hessian matrix, and parameter errors. 575 576 Arguments: 577 init_parameters : (dict) 578 the initial parameters 579 parameters : (dict) 580 the final parameters 581 min_value : (float) 582 the minimum value 583 iterations : (int) 584 the number of iterations 585 max_iterations : (int) 586 the maximum number of iterations 587 tolerance : (float) 588 the tolerance for convergence 589 convergence_flag : (int) 590 zero if the tolerance stopping criterion has been met. 591 parameters_path : (np.array) 592 the path of the minimization algorithm in parameter space 593 function_path : (np.array) 594 the path of the minimization algorithm in function space 595 hessian : (np.array) 596 the Hessian matrix of the function at the minimum 597 parameter_errors : (np.array) 598 the errors on the parameters 599 """ 600 n = self.minimizer.GetNumberOfParameters() 601 out = [( 602 self.minimizer.GetParameterName(i), 603 (self.minimizer.GetParameterValue(i), 604 self.minimizer.GetParameterScale(i)) 605 ) for i in range(n)] 606 self.results["init_parameters"] = dict(out) 607 608 self.minimizer.SetTolerance(self.tolerance) 609 self.minimizer.SetContractionRatio(self.contraction_ratio) 610 self.minimizer.SetExpansionRatio(self.expansion_ratio) 611 self.minimizer.SetMaxIterations(self.max_iterations) 612 613 self.minimizer.Minimize() 614 self.results["convergence_flag"] = not bool(self.minimizer.Iterate()) 615 616 out = [( 617 self.minimizer.GetParameterName(i), 618 self.minimizer.GetParameterValue(i), 619 ) for i in range(n)] 620 621 self.results["parameters"] = dict(out) 622 self.results["min_value"] = self.minimizer.GetFunctionValue() 623 self.results["iterations"] = self.minimizer.GetIterations() 624 self.results["max_iterations"] = self.minimizer.GetMaxIterations() 625 self.results["tolerance"] = self.minimizer.GetTolerance() 626 self.results["expansion_ratio"] = self.expansion_ratio 627 self.results["contraction_ratio"] = self.contraction_ratio 628 self.results["parameters_path"] = np.array(self.parameters_path) 629 self.results["function_path"] = np.array(self.function_path) 630 self.results["hessian"] = np.zeros((n,n)) 631 self.results["parameter_errors"] = np.zeros(n) 632 return self.results 633 634 @property 635 def x(self): 636 """Return the final parameters.""" 637 return self.results["parameters"] 638 639 @property 640 def fun(self): 641 """Return the final score value.""" 642 return self.results["min_value"] 643 644 def compute_hessian(self, epsilon=0) -> np.ndarray: 645 """ 646 Compute the Hessian matrix of `function` at the 647 minimum numerically. 648 649 Arguments: 650 epsilon : (float) 651 Step size used for numerical approximation. 652 653 Returns: 654 Hessian matrix of `function` at minimum. 655 """ 656 if not epsilon: 657 epsilon = self.tolerance * 10 658 n = self.minimizer.GetNumberOfParameters() 659 x0 = [self.minimizer.GetParameterValue(i) for i in range(n)] 660 hessian = compute_hessian(self.function, x0, epsilon=epsilon) 661 662 self.results["hessian"] = hessian 663 try: 664 ihess = np.linalg.inv(hessian) 665 cov = ihess / 2 666 self.results["parameter_errors"] = np.sqrt(np.diag(cov)) 667 print(self.results["parameter_errors"]) 668 except: 669 vedo.logger.warning("Cannot compute hessian for parameter errors") 670 self.results["parameter_errors"] = np.zeros(n) 671 return hessian 672 673 def __str__(self) -> str: 674 out = vedo.printc( 675 f"vedo.utils.Minimizer at ({hex(id(self))})".ljust(75), 676 bold=True, invert=True, return_string=True, 677 ) 678 out += "Function name".ljust(20) + self.function.__name__ + "()\n" 679 out += "-------- parameters initial value -----------\n" 680 out += "Name".ljust(20) + "Value".ljust(20) + "Scale\n" 681 for name, value in self.results["init_parameters"].items(): 682 out += name.ljust(20) + str(value[0]).ljust(20) + str(value[1]) + "\n" 683 out += "-------- parameters final value --------------\n" 684 for name, value in self.results["parameters"].items(): 685 out += name.ljust(20) + f"{value:.6f}" 686 ierr = list(self.results["parameters"]).index(name) 687 err = self.results["parameter_errors"][ierr] 688 if err: 689 out += f" ± {err:.4f}" 690 out += "\n" 691 out += "Value at minimum".ljust(20)+ f'{self.results["min_value"]}\n' 692 out += "Iterations".ljust(20) + f'{self.results["iterations"]}\n' 693 out += "Max iterations".ljust(20) + f'{self.results["max_iterations"]}\n' 694 out += "Convergence flag".ljust(20)+ f'{self.results["convergence_flag"]}\n' 695 out += "Tolerance".ljust(20) + f'{self.results["tolerance"]}\n' 696 try: 697 arr = np.array2string( 698 self.compute_hessian(), 699 separator=', ', precision=6, suppress_small=True, 700 ) 701 out += "Hessian Matrix:\n" + arr 702 except: 703 out += "Hessian Matrix: (not available)" 704 return out
A function minimizer that uses the Nelder-Mead method.
The algorithm constructs an n-dimensional simplex in parameter space (i.e. a tetrahedron if the number or parameters is 3) and moves the vertices around parameter space until a local minimum is found. The amoeba method is robust, reasonably efficient, but is not guaranteed to find the global minimum if several local minima exist.
Arguments:
- function : (callable) the function to minimize
- max_iterations : (int) the maximum number of iterations
- contraction_ratio : (float) The contraction ratio. The default value of 0.5 gives fast convergence, but larger values such as 0.6 or 0.7 provide greater stability.
- expansion_ratio : (float) The expansion ratio. The default value is 2.0, which provides rapid expansion. Values between 1.1 and 2.0 are valid.
- tol : (float) the tolerance for convergence
Example:
510 def __init__( 511 self, 512 function=None, 513 max_iterations=10000, 514 contraction_ratio=0.5, 515 expansion_ratio=2.0, 516 tol=1e-5, 517 ) -> None: 518 self.function = function 519 self.tolerance = tol 520 self.contraction_ratio = contraction_ratio 521 self.expansion_ratio = expansion_ratio 522 self.max_iterations = max_iterations 523 self.minimizer = vtki.new("AmoebaMinimizer") 524 self.minimizer.SetFunction(self._vtkfunc) 525 self.results = {} 526 self.parameters_path = [] 527 self.function_path = []
538 def eval(self, parameters=()) -> float: 539 """ 540 Evaluate the function at the current or given parameters. 541 """ 542 if len(parameters) == 0: 543 return self.minimizer.EvaluateFunction() 544 self.set_parameters(parameters) 545 return self.function(parameters)
Evaluate the function at the current or given parameters.
547 def set_parameter(self, name, value, scale=1.0) -> None: 548 """ 549 Set the parameter value. 550 The initial amount by which the parameter 551 will be modified during the search for the minimum. 552 """ 553 self.minimizer.SetParameterValue(name, value) 554 self.minimizer.SetParameterScale(name, scale)
Set the parameter value. The initial amount by which the parameter will be modified during the search for the minimum.
556 def set_parameters(self, parameters) -> None: 557 """ 558 Set the parameters names and values from a dictionary. 559 """ 560 for name, value in parameters.items(): 561 if len(value) == 2: 562 self.set_parameter(name, value[0], value[1]) 563 else: 564 self.set_parameter(name, value)
Set the parameters names and values from a dictionary.
566 def minimize(self) -> dict: 567 """ 568 Minimize the input function. 569 570 A dictionary with the minimization results 571 including the initial parameters, final parameters, 572 minimum value, number of iterations, maximum iterations, 573 tolerance, convergence flag, parameters path, 574 function path, Hessian matrix, and parameter errors. 575 576 Arguments: 577 init_parameters : (dict) 578 the initial parameters 579 parameters : (dict) 580 the final parameters 581 min_value : (float) 582 the minimum value 583 iterations : (int) 584 the number of iterations 585 max_iterations : (int) 586 the maximum number of iterations 587 tolerance : (float) 588 the tolerance for convergence 589 convergence_flag : (int) 590 zero if the tolerance stopping criterion has been met. 591 parameters_path : (np.array) 592 the path of the minimization algorithm in parameter space 593 function_path : (np.array) 594 the path of the minimization algorithm in function space 595 hessian : (np.array) 596 the Hessian matrix of the function at the minimum 597 parameter_errors : (np.array) 598 the errors on the parameters 599 """ 600 n = self.minimizer.GetNumberOfParameters() 601 out = [( 602 self.minimizer.GetParameterName(i), 603 (self.minimizer.GetParameterValue(i), 604 self.minimizer.GetParameterScale(i)) 605 ) for i in range(n)] 606 self.results["init_parameters"] = dict(out) 607 608 self.minimizer.SetTolerance(self.tolerance) 609 self.minimizer.SetContractionRatio(self.contraction_ratio) 610 self.minimizer.SetExpansionRatio(self.expansion_ratio) 611 self.minimizer.SetMaxIterations(self.max_iterations) 612 613 self.minimizer.Minimize() 614 self.results["convergence_flag"] = not bool(self.minimizer.Iterate()) 615 616 out = [( 617 self.minimizer.GetParameterName(i), 618 self.minimizer.GetParameterValue(i), 619 ) for i in range(n)] 620 621 self.results["parameters"] = dict(out) 622 self.results["min_value"] = self.minimizer.GetFunctionValue() 623 self.results["iterations"] = self.minimizer.GetIterations() 624 self.results["max_iterations"] = self.minimizer.GetMaxIterations() 625 self.results["tolerance"] = self.minimizer.GetTolerance() 626 self.results["expansion_ratio"] = self.expansion_ratio 627 self.results["contraction_ratio"] = self.contraction_ratio 628 self.results["parameters_path"] = np.array(self.parameters_path) 629 self.results["function_path"] = np.array(self.function_path) 630 self.results["hessian"] = np.zeros((n,n)) 631 self.results["parameter_errors"] = np.zeros(n) 632 return self.results
Minimize the input function.
A dictionary with the minimization results including the initial parameters, final parameters, minimum value, number of iterations, maximum iterations, tolerance, convergence flag, parameters path, function path, Hessian matrix, and parameter errors.
Arguments:
- init_parameters : (dict) the initial parameters
- parameters : (dict) the final parameters
- min_value : (float) the minimum value
- iterations : (int) the number of iterations
- max_iterations : (int) the maximum number of iterations
- tolerance : (float) the tolerance for convergence
- convergence_flag : (int) zero if the tolerance stopping criterion has been met.
- parameters_path : (np.array) the path of the minimization algorithm in parameter space
- function_path : (np.array) the path of the minimization algorithm in function space
- hessian : (np.array) the Hessian matrix of the function at the minimum
- parameter_errors : (np.array) the errors on the parameters
634 @property 635 def x(self): 636 """Return the final parameters.""" 637 return self.results["parameters"]
Return the final parameters.
639 @property 640 def fun(self): 641 """Return the final score value.""" 642 return self.results["min_value"]
Return the final score value.
644 def compute_hessian(self, epsilon=0) -> np.ndarray: 645 """ 646 Compute the Hessian matrix of `function` at the 647 minimum numerically. 648 649 Arguments: 650 epsilon : (float) 651 Step size used for numerical approximation. 652 653 Returns: 654 Hessian matrix of `function` at minimum. 655 """ 656 if not epsilon: 657 epsilon = self.tolerance * 10 658 n = self.minimizer.GetNumberOfParameters() 659 x0 = [self.minimizer.GetParameterValue(i) for i in range(n)] 660 hessian = compute_hessian(self.function, x0, epsilon=epsilon) 661 662 self.results["hessian"] = hessian 663 try: 664 ihess = np.linalg.inv(hessian) 665 cov = ihess / 2 666 self.results["parameter_errors"] = np.sqrt(np.diag(cov)) 667 print(self.results["parameter_errors"]) 668 except: 669 vedo.logger.warning("Cannot compute hessian for parameter errors") 670 self.results["parameter_errors"] = np.zeros(n) 671 return hessian
Compute the Hessian matrix of function at the
minimum numerically.
Arguments:
- epsilon : (float) Step size used for numerical approximation.
Returns:
Hessian matrix of
functionat minimum.
707def compute_hessian(func, params, bounds=None, epsilon=1e-5, verbose=True) -> np.ndarray: 708 """ 709 Compute the Hessian matrix of a scalar function `func` at `params`, 710 accounting for parameter boundaries. 711 712 Arguments: 713 func : (callable) 714 Function returning a scalar. Takes `params` as input. 715 params : (np.ndarray) 716 Parameter vector at which to compute the Hessian. 717 bounds : (list of tuples) 718 Optional bounds for parameters, e.g., [(lb1, ub1), ...]. 719 epsilon : (float) 720 Base step size for finite differences. 721 verbose : (bool) 722 Whether to print progress. 723 724 Returns: 725 Hessian matrix of shape (n_params, n_params). 726 727 Example: 728 ```python 729 from vedo import * 730 import numpy as np 731 732 # 1. Define the objective function to minimize 733 def cost_function(params): 734 a, b = params[0], params[1] 735 score = (a-5)**2 + (b-2)**2 #+a*b 736 #print(a, b, score) 737 return score 738 739 # 2. Fit parameters (example result) 740 mle_params = np.array([4.97, 1.95]) # Assume obtained via optimization 741 742 # 3. Define bounds for parameters 743 bounds = [(-np.inf, np.inf), (1e-6, 2.1)] 744 745 # 4. Compute Hessian 746 hessian = compute_hessian(cost_function, mle_params, bounds=bounds) 747 cov_matrix = np.linalg.inv(hessian) / 2 748 print("Covariance Matrix:", cov_matrix) 749 std = np.sqrt(np.diag(cov_matrix)) 750 print("Standard deviations:", std) 751 ``` 752 """ 753 n = len(params) 754 hessian = np.zeros((n, n)) 755 f_0 = func(params) # Central value 756 757 # Diagonal elements (second derivatives) 758 for i in range(n): 759 if verbose: 760 vedo.printc(f"Computing Hessian: {i+1}/{n} for diagonal", delay=0) 761 if bounds: 762 lb, ub = bounds[i] 763 else: 764 lb, ub = -np.inf, np.inf 765 766 # Adaptive step size to stay within bounds 767 h_plus = min(epsilon, ub - params[i]) # Max allowed step upward 768 h_minus = min(epsilon, params[i] - lb) # Max allowed step downward 769 h = min(h_plus, h_minus) # Use symmetric step where possible 770 771 # Avoid zero step size (if parameter is at a boundary) 772 if h <= 0: 773 h = epsilon # Fallback to one-sided derivative 774 775 # Compute f(x + h) and f(x - h) 776 params_plus = np.copy(params) 777 params_plus[i] = np.clip(params[i] + h, lb, ub) 778 f_plus = func(params_plus) 779 780 params_minus = np.copy(params) 781 params_minus[i] = np.clip(params[i] - h, lb, ub) 782 f_minus = func(params_minus) 783 784 # Central difference for diagonal 785 hessian[i, i] = (f_plus - 2*f_0 + f_minus) / (h**2) 786 787 # Off-diagonal elements (mixed partial derivatives) 788 for i in range(n): 789 if verbose: 790 print(f"Computing Hessian: {i+1}/{n} for off-diagonal ", end='') 791 for j in range(i + 1, n): 792 if verbose: 793 print(f".", end='') 794 if bounds: 795 lb_i, ub_i = bounds[i] 796 lb_j, ub_j = bounds[j] 797 else: 798 lb_i, ub_i = -np.inf, np.inf 799 lb_j, ub_j = -np.inf, np.inf 800 801 # Step sizes for i and j 802 h_i_plus = min(epsilon, ub_i - params[i]) 803 h_i_minus = min(epsilon, params[i] - lb_i) 804 h_i = min(h_i_plus, h_i_minus) 805 h_i = max(h_i, 1e-10) # Prevent division by zero 806 807 h_j_plus = min(epsilon, ub_j - params[j]) 808 h_j_minus = min(epsilon, params[j] - lb_j) 809 h_j = min(h_j_plus, h_j_minus) 810 h_j = max(h_j, 1e-10) 811 812 # Compute four perturbed points 813 params_pp = np.copy(params) 814 params_pp[i] = np.clip(params[i] + h_i, lb_i, ub_i) 815 params_pp[j] = np.clip(params[j] + h_j, lb_j, ub_j) 816 f_pp = func(params_pp) 817 818 params_pm = np.copy(params) 819 params_pm[i] = np.clip(params[i] + h_i, lb_i, ub_i) 820 params_pm[j] = np.clip(params[j] - h_j, lb_j, ub_j) 821 f_pm = func(params_pm) 822 823 params_mp = np.copy(params) 824 params_mp[i] = np.clip(params[i] - h_i, lb_i, ub_i) 825 params_mp[j] = np.clip(params[j] + h_j, lb_j, ub_j) 826 f_mp = func(params_mp) 827 828 params_mm = np.copy(params) 829 params_mm[i] = np.clip(params[i] - h_i, lb_i, ub_i) 830 params_mm[j] = np.clip(params[j] - h_j, lb_j, ub_j) 831 f_mm = func(params_mm) 832 833 # Central difference for off-diagonal 834 hessian[i, j] = (f_pp - f_pm - f_mp + f_mm) / (4 * h_i * h_j) 835 hessian[j, i] = hessian[i, j] # Symmetric 836 if verbose: 837 print() 838 return hessian
Compute the Hessian matrix of a scalar function func at params,
accounting for parameter boundaries.
Arguments:
- func : (callable)
Function returning a scalar. Takes
paramsas input. - params : (np.ndarray) Parameter vector at which to compute the Hessian.
- bounds : (list of tuples) Optional bounds for parameters, e.g., [(lb1, ub1), ...].
- epsilon : (float) Base step size for finite differences.
- verbose : (bool) Whether to print progress.
Returns:
Hessian matrix of shape (n_params, n_params).
Example:
from vedo import *
import numpy as np
# 1. Define the objective function to minimize
def cost_function(params):
a, b = params[0], params[1]
score = (a-5)**2 + (b-2)**2 #+a*b
#print(a, b, score)
return score
# 2. Fit parameters (example result)
mle_params = np.array([4.97, 1.95]) # Assume obtained via optimization
# 3. Define bounds for parameters
bounds = [(-np.inf, np.inf), (1e-6, 2.1)]
# 4. Compute Hessian
hessian = compute_hessian(cost_function, mle_params, bounds=bounds)
cov_matrix = np.linalg.inv(hessian) / 2
print("Covariance Matrix:", cov_matrix)
std = np.sqrt(np.diag(cov_matrix))
print("Standard deviations:", std)
984def geometry(obj, extent=None) -> "vedo.Mesh": 985 """ 986 Apply the `vtkGeometryFilter` to the input object. 987 This is a general-purpose filter to extract geometry (and associated data) 988 from any type of dataset. 989 This filter also may be used to convert any type of data to polygonal type. 990 The conversion process may be less than satisfactory for some 3D datasets. 991 For example, this filter will extract the outer surface of a volume 992 or structured grid dataset. 993 994 Returns a `vedo.Mesh` object. 995 996 Set `extent` as the `[xmin,xmax, ymin,ymax, zmin,zmax]` bounding box to clip data. 997 """ 998 gf = vtki.new("GeometryFilter") 999 gf.SetInputData(obj) 1000 if extent is not None: 1001 gf.SetExtent(extent) 1002 gf.Update() 1003 return vedo.Mesh(gf.GetOutput())
Apply the vtkGeometryFilter to the input object.
This is a general-purpose filter to extract geometry (and associated data)
from any type of dataset.
This filter also may be used to convert any type of data to polygonal type.
The conversion process may be less than satisfactory for some 3D datasets.
For example, this filter will extract the outer surface of a volume
or structured grid dataset.
Returns a vedo.Mesh object.
Set extent as the [xmin,xmax, ymin,ymax, zmin,zmax] bounding box to clip data.
1175def is_sequence(arg) -> bool: 1176 """Check if the input is iterable.""" 1177 if hasattr(arg, "strip"): 1178 return False 1179 if hasattr(arg, "__getslice__"): 1180 return True 1181 if hasattr(arg, "__iter__"): 1182 return True 1183 return False
Check if the input is iterable.
1549def lin_interpolate(x, rangeX, rangeY): 1550 """ 1551 Interpolate linearly the variable `x` in `rangeX` onto the new `rangeY`. 1552 If `x` is a 3D vector the linear weight is the distance to the two 3D `rangeX` vectors. 1553 1554 E.g. if `x` runs in `rangeX=[x0,x1]` and I want it to run in `rangeY=[y0,y1]` then 1555 1556 `y = lin_interpolate(x, rangeX, rangeY)` will interpolate `x` onto `rangeY`. 1557 1558 Examples: 1559 - [lin_interpolate.py](https://github.com/marcomusy/vedo/tree/master/examples/basic/lin_interpolate.py) 1560 1561  1562 """ 1563 if is_sequence(x): 1564 x = np.asarray(x) 1565 x0, x1 = np.asarray(rangeX) 1566 y0, y1 = np.asarray(rangeY) 1567 dx = x1 - x0 1568 dxn = np.linalg.norm(dx) 1569 if not dxn: 1570 return y0 1571 s = np.linalg.norm(x - x0) / dxn 1572 t = np.linalg.norm(x - x1) / dxn 1573 st = s + t 1574 out = y0 * (t / st) + y1 * (s / st) 1575 1576 else: # faster 1577 1578 x0 = rangeX[0] 1579 dx = rangeX[1] - x0 1580 if not dx: 1581 return rangeY[0] 1582 s = (x - x0) / dx 1583 out = rangeY[0] * (1 - s) + rangeY[1] * s 1584 return out
Interpolate linearly the variable x in rangeX onto the new rangeY.
If x is a 3D vector the linear weight is the distance to the two 3D rangeX vectors.
E.g. if x runs in rangeX=[x0,x1] and I want it to run in rangeY=[y0,y1] then
y = lin_interpolate(x, rangeX, rangeY) will interpolate x onto rangeY.
Examples:
1661def vector(x, y=None, z=0.0, dtype=np.float64) -> np.ndarray: 1662 """ 1663 Return a 3D numpy array representing a vector. 1664 1665 If `y` is `None`, assume input is already in the form `[x,y,z]`. 1666 """ 1667 if y is None: # assume x is already [x,y,z] 1668 return np.asarray(x, dtype=dtype) 1669 return np.array([x, y, z], dtype=dtype)
Return a 3D numpy array representing a vector.
If y is None, assume input is already in the form [x,y,z].
1680def mag(v): 1681 """Get the magnitude of a vector or array of vectors.""" 1682 v = np.asarray(v) 1683 if v.ndim == 1: 1684 return np.linalg.norm(v) 1685 return np.linalg.norm(v, axis=1)
Get the magnitude of a vector or array of vectors.
1688def mag2(v) -> np.ndarray: 1689 """Get the squared magnitude of a vector or array of vectors.""" 1690 v = np.asarray(v) 1691 if v.ndim == 1: 1692 return np.square(v).sum() 1693 return np.square(v).sum(axis=1)
Get the squared magnitude of a vector or array of vectors.
1672def versor(x, y=None, z=0.0, dtype=np.float64) -> np.ndarray: 1673 """Return the unit vector. Input can be a list of vectors.""" 1674 v = vector(x, y, z, dtype) 1675 if isinstance(v[0], np.ndarray): 1676 return np.divide(v, mag(v)[:, None]) 1677 return v / mag(v)
Return the unit vector. Input can be a list of vectors.
1762def precision(x, p: int, vrange=None, delimiter="e") -> str: 1763 """ 1764 Returns a string representation of `x` formatted to precision `p`. 1765 1766 Set `vrange` to the range in which x exists (to snap x to '0' if below precision). 1767 """ 1768 # Based on the webkit javascript implementation 1769 # `from here <https://code.google.com/p/webkit-mirror/source/browse/JavaScriptCore/kjs/number_object.cpp>`_, 1770 # and implemented by `randlet <https://github.com/randlet/to-precision>`_. 1771 # Modified for vedo by M.Musy 2020 1772 1773 if isinstance(x, str): # do nothing 1774 return x 1775 1776 if is_sequence(x): 1777 out = "(" 1778 nn = len(x) - 1 1779 for i, ix in enumerate(x): 1780 1781 try: 1782 if np.isnan(ix): 1783 return "NaN" 1784 except: 1785 # cannot handle list of list 1786 continue 1787 1788 out += precision(ix, p) 1789 if i < nn: 1790 out += ", " 1791 return out + ")" ############ <-- 1792 1793 try: 1794 if np.isnan(x): 1795 return "NaN" 1796 except TypeError: 1797 return "NaN" 1798 1799 x = float(x) 1800 1801 if x == 0.0 or (vrange is not None and abs(x) < vrange / pow(10, p)): 1802 return "0" 1803 1804 out = [] 1805 if x < 0: 1806 out.append("-") 1807 x = -x 1808 1809 e = int(np.log10(x)) 1810 # tens = np.power(10, e - p + 1) 1811 tens = 10 ** (e - p + 1) 1812 n = np.floor(x / tens) 1813 1814 # if n < np.power(10, p - 1): 1815 if n < 10 ** (p - 1): 1816 e = e - 1 1817 # tens = np.power(10, e - p + 1) 1818 tens = 10 ** (e - p + 1) 1819 n = np.floor(x / tens) 1820 1821 if abs((n + 1.0) * tens - x) <= abs(n * tens - x): 1822 n = n + 1 1823 1824 # if n >= np.power(10, p): 1825 if n >= 10 ** p: 1826 n = n / 10.0 1827 e = e + 1 1828 1829 m = "%.*g" % (p, n) 1830 if e < -2 or e >= p: 1831 out.append(m[0]) 1832 if p > 1: 1833 out.append(".") 1834 out.extend(m[1:p]) 1835 out.append(delimiter) 1836 if e > 0: 1837 out.append("+") 1838 out.append(str(e)) 1839 elif e == (p - 1): 1840 out.append(m) 1841 elif e >= 0: 1842 out.append(m[: e + 1]) 1843 if e + 1 < len(m): 1844 out.append(".") 1845 out.extend(m[e + 1 :]) 1846 else: 1847 out.append("0.") 1848 out.extend(["0"] * -(e + 1)) 1849 out.append(m) 1850 return "".join(out)
Returns a string representation of x formatted to precision p.
Set vrange to the range in which x exists (to snap x to '0' if below precision).
1715def round_to_digit(x, p) -> float: 1716 """Round a real number to the specified number of significant digits.""" 1717 if not x: 1718 return 0 1719 r = np.round(x, p - int(np.floor(np.log10(abs(x)))) - 1) 1720 if int(r) == r: 1721 return int(r) 1722 return r
Round a real number to the specified number of significant digits.
1258def point_in_triangle(p, p1, p2, p3) -> Union[bool, None]: 1259 """ 1260 Return True if a point is inside (or above/below) 1261 a triangle defined by 3 points in space. 1262 """ 1263 p1 = np.array(p1) 1264 u = p2 - p1 1265 v = p3 - p1 1266 n = np.cross(u, v) 1267 w = p - p1 1268 ln = np.dot(n, n) 1269 if not ln: 1270 return None # degenerate triangle 1271 gamma = (np.dot(np.cross(u, w), n)) / ln 1272 if 0 < gamma < 1: 1273 beta = (np.dot(np.cross(w, v), n)) / ln 1274 if 0 < beta < 1: 1275 alpha = 1 - gamma - beta 1276 if 0 < alpha < 1: 1277 return True 1278 return False
Return True if a point is inside (or above/below) a triangle defined by 3 points in space.
1471def point_line_distance(p, p1, p2) -> float: 1472 """ 1473 Compute the distance of a point to a line (not the segment) 1474 defined by `p1` and `p2`. 1475 """ 1476 return np.sqrt(vtki.vtkLine.DistanceToLine(p, p1, p2))
Compute the distance of a point to a line (not the segment)
defined by p1 and p2.
1511def closest(point, points, n=1, return_ids=False, use_tree=False): 1512 """ 1513 Returns the distances and the closest point(s) to the given set of points. 1514 Needs `scipy.spatial` library. 1515 1516 Arguments: 1517 n : (int) 1518 the nr of closest points to return 1519 return_ids : (bool) 1520 return the ids instead of the points coordinates 1521 use_tree : (bool) 1522 build a `scipy.spatial.KDTree`. 1523 An already existing one can be passed to avoid rebuilding. 1524 """ 1525 from scipy.spatial import distance, KDTree 1526 1527 points = np.asarray(points) 1528 if n == 1: 1529 dists = distance.cdist([point], points) 1530 closest_idx = np.argmin(dists) 1531 else: 1532 if use_tree: 1533 if isinstance(use_tree, KDTree): # reuse 1534 tree = use_tree 1535 else: 1536 tree = KDTree(points) 1537 dists, closest_idx = tree.query([point], k=n) 1538 closest_idx = closest_idx[0] 1539 else: 1540 dists = distance.cdist([point], points) 1541 closest_idx = np.argsort(dists)[0][:n] 1542 if return_ids: 1543 return dists, closest_idx 1544 else: 1545 return dists, points[closest_idx]
Returns the distances and the closest point(s) to the given set of points.
Needs scipy.spatial library.
Arguments:
- n : (int) the nr of closest points to return
- return_ids : (bool) return the ids instead of the points coordinates
- use_tree : (bool)
build a
scipy.spatial.KDTree. An already existing one can be passed to avoid rebuilding.
1854def grep(filename: str, tag: str, column=None, first_occurrence_only=False) -> list: 1855 """Greps the line in a file that starts with a specific `tag` string inside the file.""" 1856 import re 1857 1858 with open(str(filename), "r", encoding="UTF-8") as afile: 1859 content = [] 1860 for line in afile: 1861 if re.search(tag, line): 1862 c = line.split() 1863 c[-1] = c[-1].replace("\n", "") 1864 if column is not None: 1865 c = c[column] 1866 content.append(c) 1867 if first_occurrence_only: 1868 break 1869 return content
Greps the line in a file that starts with a specific tag string inside the file.
2152def make_bands(inputlist, n): 2153 """ 2154 Group values of a list into bands of equal value, where 2155 `n` is the number of bands, a positive integer > 2. 2156 2157 Returns a binned list of the same length as the input. 2158 """ 2159 if n < 2: 2160 return inputlist 2161 vmin = np.min(inputlist) 2162 vmax = np.max(inputlist) 2163 bb = np.linspace(vmin, vmax, n, endpoint=0) 2164 dr = bb[1] - bb[0] 2165 bb += dr / 2 2166 tol = dr / 2 * 1.001 2167 newlist = [] 2168 for s in inputlist: 2169 for b in bb: 2170 if abs(s - b) < tol: 2171 newlist.append(b) 2172 break 2173 return np.array(newlist)
Group values of a list into bands of equal value, where
n is the number of bands, a positive integer > 2.
Returns a binned list of the same length as the input.
1725def pack_spheres(bounds, radius) -> np.ndarray: 1726 """ 1727 Packing spheres into a bounding box. 1728 Returns a numpy array of sphere centers. 1729 """ 1730 h = 0.8164965 / 2 1731 d = 0.8660254 1732 a = 0.288675135 1733 1734 if is_sequence(bounds): 1735 x0, x1, y0, y1, z0, z1 = bounds 1736 else: 1737 x0, x1, y0, y1, z0, z1 = bounds.bounds() 1738 1739 x = np.arange(x0, x1, radius) 1740 nul = np.zeros_like(x) 1741 nz = int((z1 - z0) / radius / h / 2 + 1.5) 1742 ny = int((y1 - y0) / radius / d + 1.5) 1743 1744 pts = [] 1745 for iz in range(nz): 1746 z = z0 + nul + iz * h * radius 1747 dx, dy, dz = [radius * 0.5, radius * a, iz * h * radius] 1748 for iy in range(ny): 1749 y = y0 + nul + iy * d * radius 1750 if iy % 2: 1751 xs = x 1752 else: 1753 xs = x + radius * 0.5 1754 if iz % 2: 1755 p = np.c_[xs, y, z] + [dx, dy, dz] 1756 else: 1757 p = np.c_[xs, y, z] + [0, 0, dz] 1758 pts += p.tolist() 1759 return np.array(pts)
Packing spheres into a bounding box. Returns a numpy array of sphere centers.
1225def humansort(alist) -> list: 1226 """ 1227 Sort in place a given list the way humans expect. 1228 1229 E.g. `['file11', 'file1'] -> ['file1', 'file11']` 1230 1231 .. warning:: input list is modified in-place by this function. 1232 """ 1233 import re 1234 1235 def alphanum_key(s): 1236 # Turn a string into a list of string and number chunks. 1237 # e.g. "z23a" -> ["z", 23, "a"] 1238 def tryint(s): 1239 if s.isdigit(): 1240 return int(s) 1241 return s 1242 1243 return [tryint(c) for c in re.split("([0-9]+)", s)] 1244 1245 alist.sort(key=alphanum_key) 1246 return alist # NB: input list is modified
Sort in place a given list the way humans expect.
E.g. ['file11', 'file1'] -> ['file1', 'file11']
input list is modified in-place by this function.
1927def print_histogram( 1928 data, 1929 bins=10, 1930 height=10, 1931 logscale=False, 1932 minbin=0, 1933 horizontal=True, 1934 char="\U00002589", 1935 c=None, 1936 bold=True, 1937 title="histogram", 1938 spacer="", 1939) -> np.ndarray: 1940 """ 1941 Ascii histogram printing. 1942 1943 Input can be a `vedo.Volume` or `vedo.Mesh`. 1944 Returns the raw data before binning (useful when passing vtk objects). 1945 1946 Arguments: 1947 bins : (int) 1948 number of histogram bins 1949 height : (int) 1950 height of the histogram in character units 1951 logscale : (bool) 1952 use logscale for frequencies 1953 minbin : (int) 1954 ignore bins before minbin 1955 horizontal : (bool) 1956 show histogram horizontally 1957 char : (str) 1958 character to be used 1959 bold : (bool) 1960 use boldface 1961 title : (str) 1962 histogram title 1963 spacer : (str) 1964 horizontal spacer 1965 1966 Example: 1967 ```python 1968 from vedo import print_histogram 1969 import numpy as np 1970 d = np.random.normal(size=1000) 1971 data = print_histogram(d, c='b', logscale=True, title='my scalars') 1972 data = print_histogram(d, c='o') 1973 print(np.mean(data)) # data here is same as d 1974 ``` 1975  1976 """ 1977 # credits: http://pyinsci.blogspot.com/2009/10/ascii-histograms.html 1978 # adapted for vedo by M.Musy, 2019 1979 1980 if not horizontal: # better aspect ratio 1981 bins *= 2 1982 1983 try: 1984 data = vtk2numpy(data.dataset.GetPointData().GetScalars()) 1985 except AttributeError: 1986 # already an array 1987 data = np.asarray(data) 1988 1989 if isinstance(data, vtki.vtkImageData): 1990 dims = data.GetDimensions() 1991 nvx = min(100000, dims[0] * dims[1] * dims[2]) 1992 idxs = np.random.randint(0, min(dims), size=(nvx, 3)) 1993 data = [] 1994 for ix, iy, iz in idxs: 1995 d = data.GetScalarComponentAsFloat(ix, iy, iz, 0) 1996 data.append(d) 1997 data = np.array(data) 1998 1999 elif isinstance(data, vtki.vtkPolyData): 2000 arr = data.GetPointData().GetScalars() 2001 if not arr: 2002 arr = data.GetCellData().GetScalars() 2003 if not arr: 2004 return np.array([]) 2005 data = vtk2numpy(arr) 2006 2007 try: 2008 h = np.histogram(data, bins=bins) 2009 except TypeError as e: 2010 vedo.logger.error(f"cannot compute histogram: {e}") 2011 return np.array([]) 2012 2013 if minbin: 2014 hi = h[0][minbin:-1] 2015 else: 2016 hi = h[0] 2017 2018 if char == "\U00002589" and horizontal: 2019 char = "\U00002586" 2020 2021 title = title.ljust(14) + ":" 2022 entrs = " entries=" + str(len(data)) 2023 if logscale: 2024 h0 = np.log10(hi + 1) 2025 maxh0 = int(max(h0) * 100) / 100 2026 title = title + entrs + " (logscale)" 2027 else: 2028 h0 = hi 2029 maxh0 = max(h0) 2030 title = title + entrs 2031 2032 def _v(): 2033 his = "" 2034 if title: 2035 his += title + "\n" 2036 bars = h0 / maxh0 * height 2037 for l in reversed(range(1, height + 1)): 2038 line = "" 2039 if l == height: 2040 line = "%s " % maxh0 2041 else: 2042 line = " |" + " " * (len(str(maxh0)) - 3) 2043 for c in bars: 2044 if c >= np.ceil(l): 2045 line += char 2046 else: 2047 line += " " 2048 line += "\n" 2049 his += line 2050 his += "%.2f" % h[1][0] + "." * (bins) + "%.2f" % h[1][-1] + "\n" 2051 return his 2052 2053 def _h(): 2054 his = "" 2055 if title: 2056 his += title + "\n" 2057 xl = ["%.2f" % n for n in h[1]] 2058 lxl = [len(l) for l in xl] 2059 bars = h0 / maxh0 * height 2060 his += spacer + " " * int(max(bars) + 2 + max(lxl)) + "%s\n" % maxh0 2061 for i, c in enumerate(bars): 2062 line = xl[i] + " " * int(max(lxl) - lxl[i]) + "| " + char * int(c) + "\n" 2063 his += spacer + line 2064 return his 2065 2066 if horizontal: 2067 height *= 2 2068 vedo.printc(_h(), c=c, bold=bold) 2069 else: 2070 vedo.printc(_v(), c=c, bold=bold) 2071 return data
Ascii histogram printing.
Input can be a vedo.Volume or vedo.Mesh.
Returns the raw data before binning (useful when passing vtk objects).
Arguments:
- bins : (int) number of histogram bins
- height : (int) height of the histogram in character units
- logscale : (bool) use logscale for frequencies
- minbin : (int) ignore bins before minbin
- horizontal : (bool) show histogram horizontally
- char : (str) character to be used
- bold : (bool) use boldface
- title : (str) histogram title
- spacer : (str) horizontal spacer
Example:
from vedo import print_histogram import numpy as np d = np.random.normal(size=1000) data = print_histogram(d, c='b', logscale=True, title='my scalars') data = print_histogram(d, c='o') print(np.mean(data)) # data here is same as d
2128def print_inheritance_tree(C) -> None: 2129 """Prints the inheritance tree of class C.""" 2130 # Adapted from: https://stackoverflow.com/questions/26568976/ 2131 def class_tree(cls): 2132 subc = [class_tree(sub_class) for sub_class in cls.__subclasses__()] 2133 return {cls.__name__: subc} 2134 2135 def print_tree(tree, indent=8, current_ind=0): 2136 for k, v in tree.items(): 2137 if current_ind: 2138 before_dashes = current_ind - indent 2139 m = " " * before_dashes + "└" + "─" * (indent - 1) + " " + k 2140 vedo.printc(m) 2141 else: 2142 vedo.printc(k) 2143 for sub_tree in v: 2144 print_tree(sub_tree, indent=indent, current_ind=current_ind + indent) 2145 2146 if str(C.__class__) != "<class 'type'>": 2147 C = C.__class__ 2148 ct = class_tree(C) 2149 print_tree(ct)
Prints the inheritance tree of class C.
2179def camera_from_quaternion(pos, quaternion, distance=10000, ngl_correct=True) -> vtki.vtkCamera: 2180 """ 2181 Define a `vtkCamera` with a particular orientation. 2182 2183 Arguments: 2184 pos: (np.array, list, tuple) 2185 an iterator of length 3 containing the focus point of the camera 2186 quaternion: (np.array, list, tuple) 2187 a len(4) quaternion `(x,y,z,w)` describing the rotation of the camera 2188 such as returned by neuroglancer `x,y,z,w` all in `[0,1]` range 2189 distance: (float) 2190 the desired distance from pos to the camera (default = 10000 nm) 2191 2192 Returns: 2193 `vtki.vtkCamera`, a vtk camera setup according to these rules. 2194 """ 2195 camera = vtki.vtkCamera() 2196 # define the quaternion in vtk, note the swapped order 2197 # w,x,y,z instead of x,y,z,w 2198 quat_vtk = vtki.get_class("Quaternion")( 2199 quaternion[3], quaternion[0], quaternion[1], quaternion[2]) 2200 # use this to define a rotation matrix in x,y,z 2201 # right handed units 2202 M = np.zeros((3, 3), dtype=np.float32) 2203 quat_vtk.ToMatrix3x3(M) 2204 # the default camera orientation is y up 2205 up = [0, 1, 0] 2206 # calculate default camera position is backed off in positive z 2207 pos = [0, 0, distance] 2208 2209 # set the camera rototation by applying the rotation matrix 2210 camera.SetViewUp(*np.dot(M, up)) 2211 # set the camera position by applying the rotation matrix 2212 camera.SetPosition(*np.dot(M, pos)) 2213 if ngl_correct: 2214 # neuroglancer has positive y going down 2215 # so apply these azimuth and roll corrections 2216 # to fix orientatins 2217 camera.Azimuth(-180) 2218 camera.Roll(180) 2219 2220 # shift the camera posiiton and focal position 2221 # to be centered on the desired location 2222 p = camera.GetPosition() 2223 p_new = np.array(p) + pos 2224 camera.SetPosition(*p_new) 2225 camera.SetFocalPoint(*pos) 2226 return camera
Define a vtkCamera with a particular orientation.
Arguments:
- pos: (np.array, list, tuple) an iterator of length 3 containing the focus point of the camera
- quaternion: (np.array, list, tuple)
a len(4) quaternion
(x,y,z,w)describing the rotation of the camera such as returned by neuroglancerx,y,z,wall in[0,1]range - distance: (float) the desired distance from pos to the camera (default = 10000 nm)
Returns:
vtki.vtkCamera, a vtk camera setup according to these rules.
2229def camera_from_neuroglancer(state, zoom) -> vtki.vtkCamera: 2230 """ 2231 Define a `vtkCamera` from a neuroglancer state dictionary. 2232 2233 Arguments: 2234 state: (dict) 2235 an neuroglancer state dictionary. 2236 zoom: (float) 2237 how much to multiply zoom by to get camera backoff distance 2238 2239 Returns: 2240 `vtki.vtkCamera`, a vtk camera setup that matches this state. 2241 """ 2242 orient = state.get("perspectiveOrientation", [0.0, 0.0, 0.0, 1.0]) 2243 pzoom = state.get("perspectiveZoom", 10.0) 2244 position = state["navigation"]["pose"]["position"] 2245 pos_nm = np.array(position["voxelCoordinates"]) * position["voxelSize"] 2246 return camera_from_quaternion(pos_nm, orient, pzoom * zoom, ngl_correct=True)
Define a vtkCamera from a neuroglancer state dictionary.
Arguments:
- state: (dict) an neuroglancer state dictionary.
- zoom: (float) how much to multiply zoom by to get camera backoff distance
Returns:
vtki.vtkCamera, a vtk camera setup that matches this state.
2264def camera_from_dict(camera, modify_inplace=None) -> vtki.vtkCamera: 2265 """ 2266 Generate a `vtkCamera` object from a python dictionary. 2267 2268 Parameters of the camera are: 2269 - `position` or `pos` (3-tuple) 2270 - `focal_point` (3-tuple) 2271 - `viewup` (3-tuple) 2272 - `distance` (float) 2273 - `clipping_range` (2-tuple) 2274 - `parallel_scale` (float) 2275 - `thickness` (float) 2276 - `view_angle` (float) 2277 - `roll` (float) 2278 2279 Exaplanation of the parameters can be found in the 2280 [vtkCamera documentation](https://vtk.org/doc/nightly/html/classvtkCamera.html). 2281 2282 Arguments: 2283 camera: (dict) 2284 a python dictionary containing camera parameters. 2285 modify_inplace: (vtkCamera) 2286 an existing `vtkCamera` object to modify in place. 2287 2288 Returns: 2289 `vtki.vtkCamera`, a vtk camera setup that matches this state. 2290 """ 2291 if modify_inplace: 2292 vcam = modify_inplace 2293 else: 2294 vcam = vtki.vtkCamera() 2295 2296 camera = dict(camera) # make a copy so input is not emptied by pop() 2297 2298 cm_pos = camera.pop("position", camera.pop("pos", None)) 2299 cm_focal_point = camera.pop("focal_point", camera.pop("focalPoint", None)) 2300 cm_viewup = camera.pop("viewup", None) 2301 cm_distance = camera.pop("distance", None) 2302 cm_clipping_range = camera.pop("clipping_range", camera.pop("clippingRange", None)) 2303 cm_parallel_scale = camera.pop("parallel_scale", camera.pop("parallelScale", None)) 2304 cm_thickness = camera.pop("thickness", None) 2305 cm_view_angle = camera.pop("view_angle", camera.pop("viewAngle", None)) 2306 cm_roll = camera.pop("roll", None) 2307 2308 if len(camera.keys()) > 0: 2309 vedo.logger.warning(f"in camera_from_dict, key(s) not recognized: {camera.keys()}") 2310 if cm_pos is not None: vcam.SetPosition(cm_pos) 2311 if cm_focal_point is not None: vcam.SetFocalPoint(cm_focal_point) 2312 if cm_viewup is not None: vcam.SetViewUp(cm_viewup) 2313 if cm_distance is not None: vcam.SetDistance(cm_distance) 2314 if cm_clipping_range is not None: vcam.SetClippingRange(cm_clipping_range) 2315 if cm_parallel_scale is not None: vcam.SetParallelScale(cm_parallel_scale) 2316 if cm_thickness is not None: vcam.SetThickness(cm_thickness) 2317 if cm_view_angle is not None: vcam.SetViewAngle(cm_view_angle) 2318 if cm_roll is not None: vcam.SetRoll(cm_roll) 2319 return vcam
Generate a vtkCamera object from a python dictionary.
Parameters of the camera are:
positionorpos(3-tuple)focal_point(3-tuple)viewup(3-tuple)distance(float)clipping_range(2-tuple)parallel_scale(float)thickness(float)view_angle(float)roll(float)
Exaplanation of the parameters can be found in the vtkCamera documentation.
Arguments:
- camera: (dict) a python dictionary containing camera parameters.
- modify_inplace: (vtkCamera)
an existing
vtkCameraobject to modify in place.
Returns:
vtki.vtkCamera, a vtk camera setup that matches this state.
2321def camera_to_dict(vtkcam) -> dict: 2322 """ 2323 Convert a [vtkCamera](https://vtk.org/doc/nightly/html/classvtkCamera.html) 2324 object into a python dictionary. 2325 2326 Parameters of the camera are: 2327 - `position` (3-tuple) 2328 - `focal_point` (3-tuple) 2329 - `viewup` (3-tuple) 2330 - `distance` (float) 2331 - `clipping_range` (2-tuple) 2332 - `parallel_scale` (float) 2333 - `thickness` (float) 2334 - `view_angle` (float) 2335 - `roll` (float) 2336 2337 Arguments: 2338 vtkcam: (vtkCamera) 2339 a `vtkCamera` object to convert. 2340 """ 2341 cam = dict() 2342 cam["position"] = np.array(vtkcam.GetPosition()) 2343 cam["focal_point"] = np.array(vtkcam.GetFocalPoint()) 2344 cam["viewup"] = np.array(vtkcam.GetViewUp()) 2345 cam["distance"] = vtkcam.GetDistance() 2346 cam["clipping_range"] = np.array(vtkcam.GetClippingRange()) 2347 cam["parallel_scale"] = vtkcam.GetParallelScale() 2348 cam["thickness"] = vtkcam.GetThickness() 2349 cam["view_angle"] = vtkcam.GetViewAngle() 2350 cam["roll"] = vtkcam.GetRoll() 2351 return cam
Convert a vtkCamera object into a python dictionary.
Parameters of the camera are:
position(3-tuple)focal_point(3-tuple)viewup(3-tuple)distance(float)clipping_range(2-tuple)parallel_scale(float)thickness(float)view_angle(float)roll(float)
Arguments:
- vtkcam: (vtkCamera)
a
vtkCameraobject to convert.
2249def oriented_camera(center, up_vector, backoff_vector, backoff) -> vtki.vtkCamera: 2250 """ 2251 Generate a `vtkCamera` pointed at a specific location, 2252 oriented with a given up direction, set to a backoff. 2253 """ 2254 vup = np.array(up_vector) 2255 vup = vup / np.linalg.norm(vup) 2256 pt_backoff = center - backoff * np.array(backoff_vector) 2257 camera = vtki.vtkCamera() 2258 camera.SetFocalPoint(center[0], center[1], center[2]) 2259 camera.SetViewUp(vup[0], vup[1], vup[2]) 2260 camera.SetPosition(pt_backoff[0], pt_backoff[1], pt_backoff[2]) 2261 return camera
Generate a vtkCamera pointed at a specific location,
oriented with a given up direction, set to a backoff.
2577def vedo2trimesh(mesh): 2578 """ 2579 Convert `vedo.mesh.Mesh` to `Trimesh.Mesh` object. 2580 """ 2581 if is_sequence(mesh): 2582 tms = [] 2583 for a in mesh: 2584 tms.append(vedo2trimesh(a)) 2585 return tms 2586 2587 try: 2588 from trimesh import Trimesh # type: ignore 2589 except (ImportError, ModuleNotFoundError): 2590 vedo.logger.error("Need trimesh to run:\npip install trimesh") 2591 return None 2592 2593 tris = mesh.cells 2594 carr = mesh.celldata["CellIndividualColors"] 2595 ccols = carr 2596 2597 points = mesh.coordinates 2598 varr = mesh.pointdata["VertexColors"] 2599 vcols = varr 2600 2601 if len(tris) == 0: 2602 tris = None 2603 2604 return Trimesh(vertices=points, faces=tris, face_colors=ccols, vertex_colors=vcols, process=False)
Convert vedo.mesh.Mesh to Trimesh.Mesh object.
2607def trimesh2vedo(inputobj): 2608 """ 2609 Convert a `Trimesh` object to `vedo.Mesh` or `vedo.Assembly` object. 2610 """ 2611 if is_sequence(inputobj): 2612 vms = [] 2613 for ob in inputobj: 2614 vms.append(trimesh2vedo(ob)) 2615 return vms 2616 2617 inputobj_type = str(type(inputobj)) 2618 2619 if "Trimesh" in inputobj_type or "primitives" in inputobj_type: 2620 faces = inputobj.faces 2621 poly = buildPolyData(inputobj.vertices, faces) 2622 tact = vedo.Mesh(poly) 2623 if inputobj.visual.kind == "face": 2624 trim_c = inputobj.visual.face_colors 2625 elif inputobj.visual.kind == "texture": 2626 trim_c = inputobj.visual.to_color().vertex_colors 2627 else: 2628 trim_c = inputobj.visual.vertex_colors 2629 2630 if is_sequence(trim_c): 2631 if is_sequence(trim_c[0]): 2632 same_color = len(np.unique(trim_c, axis=0)) < 2 # all vtxs have same color 2633 2634 if same_color: 2635 tact.c(trim_c[0, [0, 1, 2]]).alpha(trim_c[0, 3]) 2636 else: 2637 if inputobj.visual.kind == "face": 2638 tact.cellcolors = trim_c 2639 return tact 2640 2641 if "PointCloud" in inputobj_type: 2642 2643 vdpts = vedo.shapes.Points(inputobj.vertices, r=8, c='k') 2644 if hasattr(inputobj, "vertices_color"): 2645 vcols = (inputobj.vertices_color * 1).astype(np.uint8) 2646 vdpts.pointcolors = vcols 2647 return vdpts 2648 2649 if "path" in inputobj_type: 2650 2651 lines = [] 2652 for e in inputobj.entities: 2653 # print('trimesh entity', e.to_dict()) 2654 l = vedo.shapes.Line(inputobj.vertices[e.points], c="k", lw=2) 2655 lines.append(l) 2656 return vedo.Assembly(lines) 2657 2658 return None
Convert a Trimesh object to vedo.Mesh or vedo.Assembly object.
2661def vedo2meshlab(vmesh): 2662 """Convert a `vedo.Mesh` to a Meshlab object.""" 2663 try: 2664 import pymeshlab as mlab # type: ignore 2665 except ModuleNotFoundError: 2666 vedo.logger.error("Need pymeshlab to run:\npip install pymeshlab") 2667 2668 vertex_matrix = vmesh.vertices.astype(np.float64) 2669 2670 try: 2671 face_matrix = np.asarray(vmesh.cells, dtype=np.float64) 2672 except: 2673 print("WARNING: in vedo2meshlab(), need to triangulate mesh first!") 2674 face_matrix = np.array(vmesh.clone().triangulate().cells, dtype=np.float64) 2675 2676 # v_normals_matrix = vmesh.normals(cells=False, recompute=False) 2677 v_normals_matrix = vmesh.vertex_normals 2678 if not v_normals_matrix.shape[0]: 2679 v_normals_matrix = np.empty((0, 3), dtype=np.float64) 2680 2681 # f_normals_matrix = vmesh.normals(cells=True, recompute=False) 2682 f_normals_matrix = vmesh.cell_normals 2683 if not f_normals_matrix.shape[0]: 2684 f_normals_matrix = np.empty((0, 3), dtype=np.float64) 2685 2686 v_color_matrix = vmesh.pointdata["RGBA"] 2687 if v_color_matrix is None: 2688 v_color_matrix = np.empty((0, 4), dtype=np.float64) 2689 else: 2690 v_color_matrix = v_color_matrix.astype(np.float64) / 255 2691 if v_color_matrix.shape[1] == 3: 2692 v_color_matrix = np.c_[ 2693 v_color_matrix, np.ones(v_color_matrix.shape[0], dtype=np.float64) 2694 ] 2695 2696 f_color_matrix = vmesh.celldata["RGBA"] 2697 if f_color_matrix is None: 2698 f_color_matrix = np.empty((0, 4), dtype=np.float64) 2699 else: 2700 f_color_matrix = f_color_matrix.astype(np.float64) / 255 2701 if f_color_matrix.shape[1] == 3: 2702 f_color_matrix = np.c_[ 2703 f_color_matrix, np.ones(f_color_matrix.shape[0], dtype=np.float64) 2704 ] 2705 2706 m = mlab.Mesh( 2707 vertex_matrix=vertex_matrix, 2708 face_matrix=face_matrix, 2709 v_normals_matrix=v_normals_matrix, 2710 f_normals_matrix=f_normals_matrix, 2711 v_color_matrix=v_color_matrix, 2712 f_color_matrix=f_color_matrix, 2713 ) 2714 2715 for k in vmesh.pointdata.keys(): 2716 data = vmesh.pointdata[k] 2717 if data is not None: 2718 if data.ndim == 1: # scalar 2719 m.add_vertex_custom_scalar_attribute(data.astype(np.float64), k) 2720 elif data.ndim == 2: # vectorial data 2721 if "tcoord" not in k.lower() and k not in ["Normals", "TextureCoordinates"]: 2722 m.add_vertex_custom_point_attribute(data.astype(np.float64), k) 2723 2724 for k in vmesh.celldata.keys(): 2725 data = vmesh.celldata[k] 2726 if data is not None: 2727 if data.ndim == 1: # scalar 2728 m.add_face_custom_scalar_attribute(data.astype(np.float64), k) 2729 elif data.ndim == 2 and k != "Normals": # vectorial data 2730 m.add_face_custom_point_attribute(data.astype(np.float64), k) 2731 2732 m.update_bounding_box() 2733 return m
Convert a vedo.Mesh to a Meshlab object.
2736def meshlab2vedo(mmesh, pointdata_keys=(), celldata_keys=()): 2737 """Convert a Meshlab object to `vedo.Mesh`.""" 2738 inputtype = str(type(mmesh)) 2739 2740 if "MeshSet" in inputtype: 2741 mmesh = mmesh.current_mesh() 2742 2743 mpoints, mcells = mmesh.vertex_matrix(), mmesh.face_matrix() 2744 if len(mcells) > 0: 2745 polydata = buildPolyData(mpoints, mcells) 2746 else: 2747 polydata = buildPolyData(mpoints, None) 2748 2749 if mmesh.has_vertex_scalar(): 2750 parr = mmesh.vertex_scalar_array() 2751 parr_vtk = numpy_to_vtk(parr) 2752 parr_vtk.SetName("MeshLabScalars") 2753 polydata.GetPointData().AddArray(parr_vtk) 2754 polydata.GetPointData().SetActiveScalars("MeshLabScalars") 2755 2756 if mmesh.has_face_scalar(): 2757 carr = mmesh.face_scalar_array() 2758 carr_vtk = numpy_to_vtk(carr) 2759 carr_vtk.SetName("MeshLabScalars") 2760 polydata.GetCellData().AddArray(carr_vtk) 2761 polydata.GetCellData().SetActiveScalars("MeshLabScalars") 2762 2763 for k in pointdata_keys: 2764 parr = mmesh.vertex_custom_scalar_attribute_array(k) 2765 parr_vtk = numpy_to_vtk(parr) 2766 parr_vtk.SetName(k) 2767 polydata.GetPointData().AddArray(parr_vtk) 2768 polydata.GetPointData().SetActiveScalars(k) 2769 2770 for k in celldata_keys: 2771 carr = mmesh.face_custom_scalar_attribute_array(k) 2772 carr_vtk = numpy_to_vtk(carr) 2773 carr_vtk.SetName(k) 2774 polydata.GetCellData().AddArray(carr_vtk) 2775 polydata.GetCellData().SetActiveScalars(k) 2776 2777 pnorms = mmesh.vertex_normal_matrix() 2778 if len(pnorms) > 0: 2779 polydata.GetPointData().SetNormals(numpy2vtk(pnorms, name="Normals")) 2780 2781 cnorms = mmesh.face_normal_matrix() 2782 if len(cnorms) > 0: 2783 polydata.GetCellData().SetNormals(numpy2vtk(cnorms, name="Normals")) 2784 return vedo.Mesh(polydata)
Convert a Meshlab object to vedo.Mesh.
2795def vedo2open3d(vedo_mesh): 2796 """ 2797 Return an `open3d.geometry.TriangleMesh` version of the current mesh. 2798 """ 2799 try: 2800 import open3d as o3d # type: ignore 2801 except RuntimeError: 2802 vedo.logger.error("Need open3d to run:\npip install open3d") 2803 2804 # create from numpy arrays 2805 o3d_mesh = o3d.geometry.TriangleMesh( 2806 vertices=o3d.utility.Vector3dVector(vedo_mesh.vertices), 2807 triangles=o3d.utility.Vector3iVector(vedo_mesh.cells), 2808 ) 2809 # TODO: need to add some if check here in case color and normals 2810 # info are not existing 2811 # o3d_mesh.vertex_colors = o3d.utility.Vector3dVector(vedo_mesh.pointdata["RGB"]/255) 2812 # o3d_mesh.vertex_normals= o3d.utility.Vector3dVector(vedo_mesh.pointdata["Normals"]) 2813 return o3d_mesh
Return an open3d.geometry.TriangleMesh version of the current mesh.
2787def open3d2vedo(o3d_mesh): 2788 """Convert `open3d.geometry.TriangleMesh` to a `vedo.Mesh`.""" 2789 m = vedo.Mesh([np.array(o3d_mesh.vertices), np.array(o3d_mesh.triangles)]) 2790 # TODO: could also check whether normals and color are present in 2791 # order to port with the above vertices/faces 2792 return m
Convert open3d.geometry.TriangleMesh to a vedo.Mesh.
926def vtk2numpy(varr): 927 """Convert a `vtkDataArray`, `vtkIdList` or `vtTransform` into a numpy array.""" 928 if varr is None: 929 return np.array([]) 930 if isinstance(varr, vtki.vtkIdList): 931 return np.array([varr.GetId(i) for i in range(varr.GetNumberOfIds())]) 932 elif isinstance(varr, vtki.vtkBitArray): 933 carr = vtki.vtkCharArray() 934 carr.DeepCopy(varr) 935 varr = carr 936 elif isinstance(varr, vtki.vtkHomogeneousTransform): 937 try: 938 varr = varr.GetMatrix() 939 except AttributeError: 940 pass 941 M = [[varr.GetElement(i, j) for j in range(4)] for i in range(4)] 942 return np.array(M) 943 return vtk_to_numpy(varr)
Convert a vtkDataArray, vtkIdList or vtTransform into a numpy array.
897def numpy2vtk(arr, dtype=None, deep=True, name=""): 898 """ 899 Convert a numpy array into a `vtkDataArray`. 900 Use `dtype='id'` for `vtkIdTypeArray` objects. 901 """ 902 # https://github.com/Kitware/VTK/blob/master/Wrapping/Python/vtkmodules/util/numpy_support.py 903 if arr is None: 904 return None 905 906 arr = np.ascontiguousarray(arr) 907 908 if dtype == "id": 909 if vtki.vtkIdTypeArray().GetDataTypeSize() != 4: 910 ast = np.int64 911 else: 912 ast = np.int32 913 varr = numpy_to_vtkIdTypeArray(arr.astype(ast), deep=deep) 914 elif dtype: 915 varr = numpy_to_vtk(arr.astype(dtype), deep=deep) 916 else: 917 # let numpy_to_vtk() decide what is best type based on arr type 918 if arr.dtype == np.bool_: 919 arr = arr.astype(np.uint8) 920 varr = numpy_to_vtk(arr, deep=deep) 921 922 if name: 923 varr.SetName(name) 924 return varr
Convert a numpy array into a vtkDataArray.
Use dtype='id' for vtkIdTypeArray objects.
1587def get_uv(p, x, v): 1588 """ 1589 Obtain the texture uv-coords of a point p belonging to a face that has point 1590 coordinates (x0, x1, x2) with the corresponding uv-coordinates v=(v0, v1, v2). 1591 All p and x0,x1,x2 are 3D-vectors, while v are their 2D uv-coordinates. 1592 1593 Example: 1594 ```python 1595 from vedo import * 1596 1597 pic = Image(dataurl+"coloured_cube_faces.jpg") 1598 cb = Mesh(dataurl+"coloured_cube.obj").lighting("off").texture(pic) 1599 1600 cbpts = cb.points 1601 faces = cb.cells 1602 uv = cb.pointdata["Material"] 1603 1604 pt = [-0.2, 0.75, 2] 1605 pr = cb.closest_point(pt) 1606 1607 idface = cb.closest_point(pt, return_cell_id=True) 1608 idpts = faces[idface] 1609 uv_face = uv[idpts] 1610 1611 uv_pr = utils.get_uv(pr, cbpts[idpts], uv_face) 1612 print("interpolated uv =", uv_pr) 1613 1614 sx, sy = pic.dimensions() 1615 i_interp_uv = uv_pr * [sy, sx] 1616 ix, iy = i_interp_uv.astype(int) 1617 mpic = pic.tomesh() 1618 rgba = mpic.pointdata["RGBA"].reshape(sy, sx, 3) 1619 print("color =", rgba[ix, iy]) 1620 1621 show( 1622 [[cb, Point(pr), cb.labels("Material")], 1623 [pic, Point(i_interp_uv)]], 1624 N=2, axes=1, sharecam=False, 1625 ).close() 1626 ``` 1627  1628 """ 1629 # Vector vp=p-x0 is representable as alpha*s + beta*t, 1630 # where s = x1-x0 and t = x2-x0, in matrix form 1631 # vp = [alpha, beta] . matrix(s,t) 1632 # M = matrix(s,t) is 2x3 matrix, so (alpha, beta) can be found by 1633 # inverting any of its minor A with non-zero determinant. 1634 # Once found, uv-coords of p are vt0 + alpha (vt1-v0) + beta (vt2-v0) 1635 1636 p = np.asarray(p) 1637 x0, x1, x2 = np.asarray(x)[:3] 1638 vt0, vt1, vt2 = np.asarray(v)[:3] 1639 1640 s = x1 - x0 1641 t = x2 - x0 1642 vs = vt1 - vt0 1643 vt = vt2 - vt0 1644 vp = p - x0 1645 1646 # finding a minor with independent rows 1647 M = np.matrix([s, t]) 1648 mnr = [0, 1] 1649 A = M[:, mnr] 1650 if np.abs(np.linalg.det(A)) < 0.000001: 1651 mnr = [0, 2] 1652 A = M[:, mnr] 1653 if np.abs(np.linalg.det(A)) < 0.000001: 1654 mnr = [1, 2] 1655 A = M[:, mnr] 1656 Ainv = np.linalg.inv(A) 1657 alpha_beta = vp[mnr].dot(Ainv) # [alpha, beta] 1658 return np.asarray(vt0 + alpha_beta.dot(np.matrix([vs, vt])))[0]
Obtain the texture uv-coords of a point p belonging to a face that has point coordinates (x0, x1, x2) with the corresponding uv-coordinates v=(v0, v1, v2). All p and x0,x1,x2 are 3D-vectors, while v are their 2D uv-coordinates.
Example:
from vedo import * pic = Image(dataurl+"coloured_cube_faces.jpg") cb = Mesh(dataurl+"coloured_cube.obj").lighting("off").texture(pic) cbpts = cb.points faces = cb.cells uv = cb.pointdata["Material"] pt = [-0.2, 0.75, 2] pr = cb.closest_point(pt) idface = cb.closest_point(pt, return_cell_id=True) idpts = faces[idface] uv_face = uv[idpts] uv_pr = utils.get_uv(pr, cbpts[idpts], uv_face) print("interpolated uv =", uv_pr) sx, sy = pic.dimensions() i_interp_uv = uv_pr * [sy, sx] ix, iy = i_interp_uv.astype(int) mpic = pic.tomesh() rgba = mpic.pointdata["RGBA"].reshape(sy, sx, 3) print("color =", rgba[ix, iy]) show( [[cb, Point(pr), cb.labels("Material")], [pic, Point(i_interp_uv)]], N=2, axes=1, sharecam=False, ).close()
842def andrews_curves(M, res=100) -> np.ndarray: 843 """ 844 Computes the [Andrews curves](https://en.wikipedia.org/wiki/Andrews_plot) 845 for the provided data. 846 847 The input array is an array of shape (n,m) where n is the number of 848 features and m is the number of observations. 849 850 Arguments: 851 M : (ndarray) 852 the data matrix (or data vector). 853 res : (int) 854 the resolution (n. of points) of the output curve. 855 856 Example: 857 - [andrews_cluster.py](https://github.com/marcomusy/vedo/blob/master/examples/pyplot/andrews_cluster.py) 858 859  860 """ 861 # Credits: 862 # https://gist.github.com/ryuzakyl/12c221ff0e54d8b1ac171c69ea552c0a 863 M = np.asarray(M) 864 m = int(res + 0.5) 865 866 # getting data vectors 867 X = np.reshape(M, (1, -1)) if len(M.shape) == 1 else M.copy() 868 _rows, n = X.shape 869 870 # andrews curve dimension (n. theta angles) 871 t = np.linspace(-np.pi, np.pi, m) 872 873 # m: range of values for angle theta 874 # n: amount of components of the Fourier expansion 875 A = np.empty((m, n)) 876 877 # setting first column of A 878 A[:, 0] = [1/np.sqrt(2)] * m 879 880 # filling columns of A 881 for i in range(1, n): 882 # computing the scaling coefficient for angle theta 883 c = np.ceil(i / 2) 884 # computing i-th column of matrix A 885 col = np.sin(c * t) if i % 2 == 1 else np.cos(c * t) 886 # setting column in matrix A 887 A[:, i] = col[:] 888 889 # computing Andrews curves for provided data 890 andrew_curves = np.dot(A, X.T).T 891 892 # returning the Andrews Curves (raveling if needed) 893 return np.ravel(andrew_curves) if andrew_curves.shape[0] == 1 else andrew_curves
Computes the Andrews curves for the provided data.
The input array is an array of shape (n,m) where n is the number of features and m is the number of observations.
Arguments:
- M : (ndarray) the data matrix (or data vector).
- res : (int) the resolution (n. of points) of the output curve.





